[Project] Forest Classification
1. Project Introduction
GOAL
이번에 Data Science Lab에서 진행하게 된 프로젝트는 “Forest Classification”,즉 숲의 유형(종류)를 구분하는 것이었다. 이 데이터는 Roosevelt National Forest of northern Colorado 지역을 대상으로 하였고, 우리가 구분하고자 했던 숲의 종류 7가지는 다음과 같다 :
- Spruce/Fir
- Lodgepole Pine
- Ponderosa Pine
- Cottonwood/Willow
- Aspen
- Douglas-fir
- Krummholz
DATA INTRODUCTION
Training Dataset은 총 15,120개이고, Test Dataset은 그 보다 훨씬 많은 565,892개다. 숲의 유형을 예측하는데 사용할 변수들은 다음과 같았다.
- Elevation - Elevation in meters
- Aspect - Aspect in degrees azimuth
- Slope - Slope in degrees
- Horizontal_Distance_To_Hydrology - Horz Dist to nearest surface water features
- Vertical_Distance_To_Hydrology - Vert Dist to nearest surface water features
- Horizontal_Distance_To_Roadways - Horz Dist to nearest roadway
- Hillshade_9am (0 to 255 index) - Hillshade index at 9am, summer solstice
- Hillshade_Noon (0 to 255 index) - Hillshade index at noon, summer solstice
- Hillshade_3pm (0 to 255 index) - Hillshade index at 3pm, summer solstice
- Horizontal_Distance_To_Fire_Points - Horz Dist to nearest wildfire ignition points
- Wilderness_Area (4 binary columns, 0 = absence or 1 = presence) - Wilderness area designation
- Soil_Type (40 binary columns, 0 = absence or 1 = presence) - Soil Type designation
- Cover_Type (7 types, integers 1 to 7) - Forest Cover Type designation
위의 변수에서 카테고리 변수인 “11.Wilderness_Area”의 4 종류는 다음 중 하나이다.
- 1) Rawah Wilderness Area
- 2) Neota Wilderness Area
- 3) Comanche Peak Wilderness Area
- 4) Cache la Poudre Wilderness Area
위의 변수에서 카테고리 변수인 “12.Soil_Type”의 40 종류는 다음 중 하나이다. (생략)
The soil types are:
- 1) Cathedral family - Rock outcrop complex, extremely stony.
- 2) Vanet - Ratake families complex, very stony …
- 39) Moran family - Cryorthents - Leighcan family complex, extremely stony.
- 40) Moran family - Cryorthents - Rock land complex, extremely stony.
2. Packages & Data
Packages
Ensemble 모델들을 사용하여 분류를 할 것이다.
( 대표 모델 : Random Forest, XGBoost, LightGBM )
# Visualization & basic packages
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.gridspec as gridspec
import itertools
import pandas_profiling
from mlxtend.plotting import plot_learning_curves
from mlxtend.plotting import plot_decision_regions
%matplotlib inline
# Modeling
from sklearn import datasets
from statsmodels import api as sm # For poisson Regression
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, BaggingClassifier
from sklearn.ensemble import RandomForestRegressor
import xgboost as xgb
import lightgbm as lgb
from sklearn.linear_model import LogisticRegression
# Evaluation
from sklearn.model_selection import cross_val_score, train_test_split
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV
import warnings
warnings.filterwarnings('ignore')
Data
train = pd.read_csv("train.csv")
test = pd.read_csv("test.csv")
train.shape, test.shape
((15120, 56), (565892, 55))
- 예측해야하는 종속 변수 : “Cover_Type” (숲의 종류)
set(train.columns.values) - set(test.columns.values)
{'Cover_Type'}
Pandas Profiling을 통한 data 요약
-
1) 숲 종류 1~7이 고르게 (2160개 씩)있는 data다
-
2) 토양 종류 40종류 너무 많다 (토양7,15는 없다 -> but test data에는 있을수도 있으니 drop X )
-
3) 야생 지역 4종류 있다.
-
4) NA값 없다!
3. EDA
Q1. 야생 지역 (Wilderness_Area)에 따른 Cover_Type(Y값)은?
# Id drop
train.drop('Id', axis=1, inplace=True)
# boolean -> float
train.iloc[:,-45:] = train.iloc[:,-45:].astype(float)
train.columns[-45:-41]
Index(['Wilderness_Area1', 'Wilderness_Area2', 'Wilderness_Area3',
'Wilderness_Area4'],
dtype='object')
(1) Graph
- 아래 그래프를 통해서 알 수 있 듯, 숲의 종류 마다 ‘wilderness area’가 매우 다름을 확인할 수 있다. 아직 모델링을 하지 않았지만, 뭔가 classification을 하는 데에 있어서 중요한 변수로 작용할 수 있다는 생각이 든다.
for i in train.columns[-45:-41]:
plt.figure()
ratio = pd.crosstab(train[i], train.Cover_Type, normalize='index').iloc[1,:].round(3)
ratio.plot(kind='bar')
plt.title(i, fontsize=20)
plt.show()
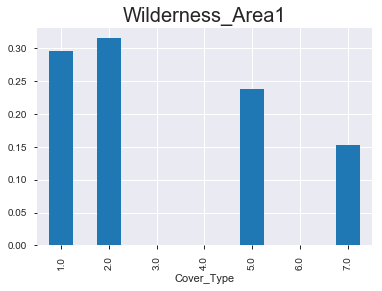
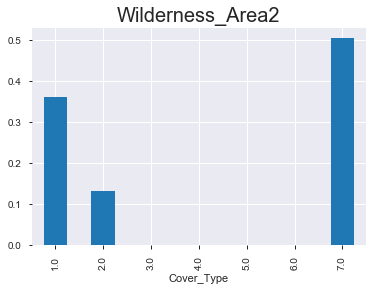
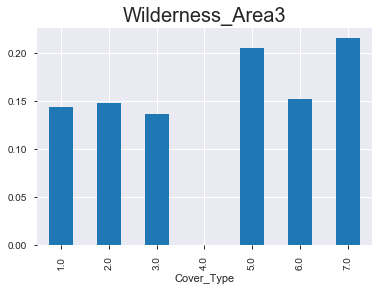
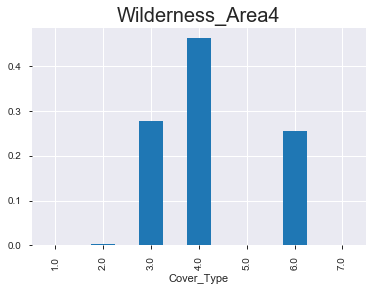
(2) 상대적 비율 & 절대적 값
- 위의 그래프를 수치로 확인해보면 다음과 같다.
for i in train.columns[-45:-41]:
ratio = pd.crosstab(train[i], train.Cover_Type, normalize='index').iloc[1,:].round(3) # Cover_Type의 상대적 값 (비율)
value = pd.crosstab(train[i], train.Cover_Type).iloc[1,:] # Cover_Type의 절대적 값 (개수)
print("<",i,">")
print(pd.concat([ratio,value],axis=1),'\n\n')
< Wilderness_Area1 >
1.0 1.0
Cover_Type
1.0 0.295 1062
2.0 0.315 1134
3.0 0.000 0
4.0 0.000 0
5.0 0.238 856
6.0 0.000 0
7.0 0.152 545
< Wilderness_Area2 >
1.0 1.0
Cover_Type
1.0 0.363 181
2.0 0.132 66
3.0 0.000 0
4.0 0.000 0
5.0 0.000 0
6.0 0.000 0
7.0 0.505 252
< Wilderness_Area3 >
1.0 1.0
Cover_Type
1.0 0.144 917
2.0 0.148 940
3.0 0.136 863
4.0 0.000 0
5.0 0.205 1304
6.0 0.152 962
7.0 0.215 1363
< Wilderness_Area4 >
1.0 1.0
Cover_Type
1.0 0.000 0
2.0 0.004 20
3.0 0.277 1297
4.0 0.462 2160
5.0 0.000 0
6.0 0.256 1198
7.0 0.000 0
Q2. 토양 종류 (Soil_Type)에 따른 Cover_Type(Y값)은?
(1) Graph
for i in train.columns[-41:-1]:
if (i != 'Soil_Type7') & (i!='Soil_Type15'):
plt.figure()
ratio = pd.crosstab(train[i], train.Cover_Type, normalize='index').iloc[1,:].round(3)
ratio.plot(kind='bar')
plt.title(i, fontsize=20)
plt.show()
(2) 상대적 비율 & 절대적 값
for i in train.columns[-41:-1]:
if (i != 'Soil_Type7') & (i!='Soil_Type15'):
ratio = pd.crosstab(train[i], train.Cover_Type, normalize='index').iloc[1,:].round(3)
value = pd.crosstab(train[i], train.Cover_Type).iloc[1,:]
print("<",i,">")
print(pd.concat([ratio,value],axis=1),'\n\n')
Q3. 그 외의 Numerical 변수들에 따른 Cover_Type(Y값)은?
for i in train.columns[0:9]:
plt.figure()
sns.boxplot(x=train.Cover_Type, y = train[i])
plt.title(i, fontsize=20)
plt.show()
4. Modeling
- 사용할 4개의 모델 :
1) Decision Tree
2) Random Forest
3) LightGBM
4) XGBoost
Train = train.iloc[:,:-1]
Test = train.iloc[:,-1]
x_train,x_test,y_train,y_test = train_test_split(Train,Test, test_size=0.2,random_state=42)
x_train = x_train.reset_index(drop=True)
x_test = x_test.reset_index(drop=True)
y_train = y_train.reset_index(drop=True)
y_test = y_test.reset_index(drop=True)
X = x_train
y = y_train
5-fold CV를 통해, validation dataset의 accuracy가 가장 높은 모델을 선택할 것이다.
Default
- hyperparameter tuning 이전
- 최고 성능을 보인 것은 LightGBM : 84.38%이었다.
names = ['DecisionTree', 'RandomForest', 'LGBM', 'XGB']
clf_list = [DecisionTreeClassifier(random_state=42), RandomForestClassifier(random_state=42), lgb.LGBMClassifier(random_state=42), xgb.XGBClassifier(objective = 'multi:softprob',random_state=42) ]
for name, clf in zip(names, clf_list):
clf.fit(X,y)
print('---- {} ----'.format(name))
print('cv score : ', cross_val_score(clf, X, y, cv=5).mean())
---- DecisionTree ----
cv score : 0.7683527742073142
---- RandomForest ----
cv score : 0.8197792619059616
---- LGBM ----
cv score : 0.8438330690017872
---- XGB ----
cv score : 0.7504086609575861
Hyperparameter Tuning
- 최적의 hyperparameter를 찾기 위해, 두 가지 방식 (Grid Search & Random Search)을 사용하였다.
from scipy.stats import randint
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV
def hypertuning_rscv(est, p_distr, nbr_iter,X,y):
rdmsearch = RandomizedSearchCV(est, param_distributions=p_distr, n_jobs=-1, n_iter=nbr_iter, cv=5, random_state=0)
#CV = Cross-Validation ( here using Stratified KFold CV)
rdmsearch.fit(X,y)
ht_params = rdmsearch.best_params_
ht_score = rdmsearch.best_score_
return ht_params, ht_score
def hypertuning_gscv(est, p_distr,X,y):
gdsearch = GridSearchCV(est, param_grid=p_distr, n_jobs=-1, cv=5)
gdsearch.fit(X,y)
bt_param = gdsearch.best_params_
bt_score = gdsearch.best_score_
return bt_param, bt_score
dt_params = {"criterion": ["gini", "entropy"],
"min_samples_split": randint(2, 20),
"max_depth": randint(1, 20),
"min_samples_leaf": randint(1, 20),
"max_leaf_nodes": randint(2, 20)}
rf_params = {'max_depth':np.arange(3, 30),
'n_estimators':np.arange(100, 400),
'min_samples_split':np.arange(2, 10)}
lgbm_params ={'max_depth': np.arange(3, 30),
'num_leaves': np.arange(10, 100),
'learning_rate': [ 0.01, 0.05, 0.01, 0.001],
'min_child_samples': randint(2, 30),
'min_child_weight': [1e-5, 1e-3, 1e-2, 1e-1, 1, 1e1, 1e2, 1e3, 1e4],
'subsample': np.linspace(0.6, 0.9, 30, endpoint=True),
'colsample_bytree': np.linspace(0.1, 0.8, 100, endpoint=True),
'reg_alpha': [0, 1e-1, 1, 2, 5, 7, 10, 50, 100],
'reg_lambda': [0, 1e-1, 1, 5, 10, 20, 50, 100],
'n_estimators': np.arange(100, 400)}
xgb_params = {'eta': np.linspace(0.001, 0.4, 50),
'min_child_weight': [1, 5, 10],
'gamma': np.arange(0, 20),
'subsample': [0.6, 0.8, 1.0],
'colsample_bytree': [0.6, 0.8, 1.0],
'max_depth': np.arange(1, 500)}
params_list = [dt_params, rf_params, lgbm_params, xgb_params]
각 모델 별로 최적의 hyperparameter값을 best_param_dict에 넣는다.
best_param_dict = dict()
print('5-fold cross validation scores & best parameters :\n')
for name, clf, param_list in zip(names, clf_list, params_list):
print('---- {} with RandomSearch ----'.format(name))
best_params = hypertuning_rscv(clf, param_list, 30, X, y)
best_param_dict[name] = best_params[0]
print('best_params : ', best_params[0])
clf.set_params(**best_params[0])
cv_score = cross_val_score(clf, X, y, cv=5).mean()
print('cv score : ', cv_score)
그 결과는 아래와 같다.
- 이번에는 RandomForest가 84.95%로 가장 좋은 성능을 보였다.
- 최적의 hyperparameter :
- n_estimator (분류기의 개수) : 391
- min_samples_split (분기가 일어나기 위한 최소한의 sample 개수) : 2
- max_depth (tree의 최대 깊이) : 22
5-fold cross validation scores & best parameters :
---- DecisionTree with RandomSearch ----
best_params : {'criterion': 'gini', 'max_depth': 18, 'max_leaf_nodes': 17, 'min_samples_leaf': 5, 'min_samples_split': 11}
cv score : 0.647404544674101
---- RandomForest with RandomSearch ----
best_params : {'n_estimators': 391, 'min_samples_split': 2, 'max_depth': 22}
cv score : 0.8495366799306494
---- LGBM with RandomSearch ----
best_params : {'colsample_bytree': 0.4747474747474748, 'learning_rate': 0.05, 'max_depth': 9, 'min_child_samples': 10, 'min_child_weight': 1, 'n_estimators': 373, 'num_leaves': 89, 'reg_alpha': 5, 'reg_lambda': 0.1, 'subsample': 0.7034482758620689}
cv score : 0.8291166436732764
---- XGB with RandomSearch ----
best_params : {'subsample': 0.6, 'min_child_weight': 5, 'max_depth': 356, 'gamma': 0, 'eta': 0.20457142857142857, 'colsample_bytree': 0.8}
cv score : 0.8467260432797161
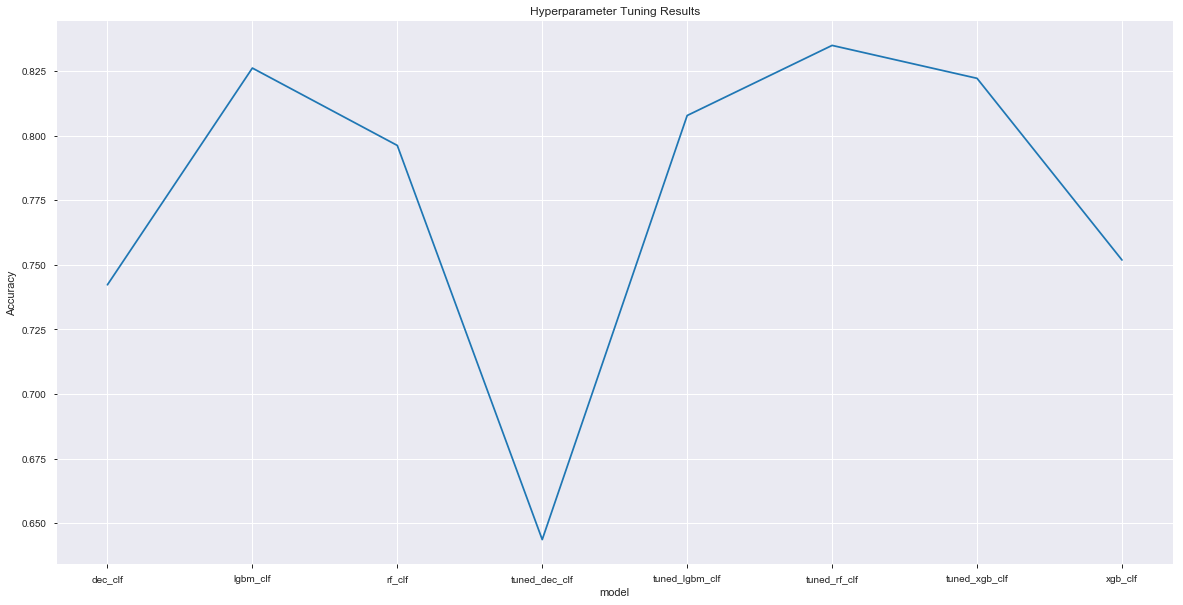
총 8개의 모델의 성능을 보면 다음과 같다. (tuning 이전 4개 + tuning 이후 4)
names = ['dec_clf', 'rf_clf','lgbm_clf','xgb_clf','tuned_dec_clf','tuned_rf_clf','tuned_lgbm_clf',
'tuned_xgb_clf']
labels = ['DecisionTree', 'RandomForest', 'LGBM','XGB',
'Tuned_DecisionTree', 'Tuned_RandomForest', 'Tuned_LGBM','Tuned_XGB']
clf_list = [DecisionTreeClassifier(random_state=42),
RandomForestClassifier(random_state=42),
lgb.LGBMClassifier(random_state=42),
xgb.XGBClassifier(random_state=42),
DecisionTreeClassifier(random_state=42, **best_param_dict['DecisionTree']),
RandomForestClassifier(random_state=42, **best_param_dict['RandomForest']),
lgb.LGBMClassifier(random_state=42, **best_param_dict['LGBM']),
xgb.XGBClassifier(random_state=42, **best_param_dict['XGB'])]
scores_list = dict()
for name, label, clf in zip(names, labels, clf_list):
print('---- {} ----'.format(name))
scores = cross_val_score(clf, X, y, cv=2, scoring='accuracy')
scores_list[name] = scores[0]
print ("Accuracy: %.2f (+/- %.2f) [%s]" %(scores.mean(), scores.std(), label))
plt.figure(figsize=(20,10))
plt.plot(*zip(*sorted(scores_list.items())))
plt.ylabel('Accuracy'); plt.xlabel('model'); plt.title('Hyperparameter Tuning Results');
plt.show()
---- dec_clf ----
Accuracy: 0.74 (+/- 0.00) [DecisionTree]
---- rf_clf ----
Accuracy: 0.79 (+/- 0.01) [RandomForest]
---- lgbm_clf ----
Accuracy: 0.82 (+/- 0.00) [LGBM]
---- xgb_clf ----
Accuracy: 0.75 (+/- 0.01) [XGB]
---- tuned_dec_clf ----
Accuracy: 0.65 (+/- 0.00) [Tuned_DecisionTree]
---- tuned_rf_clf ----
Accuracy: 0.83 (+/- 0.01) [Tuned_RandomForest]
---- tuned_lgbm_clf ----
Accuracy: 0.80 (+/- 0.01) [Tuned_LGBM]
---- tuned_xgb_clf ----
Accuracy: 0.82 (+/- 0.01) [Tuned_XGB]
Stacking
지금 까지 만든 모델들을 사용해서 Stacking을 해보았다. Meta Learner로는 Logistic Regression을 사용하였다.
from mlxtend.classifier import StackingCVClassifier
from sklearn.linear_model import LinearRegression, LogisticRegression
lr = LogisticRegression()
ens_dt = BaggingClassifier(DecisionTreeClassifier(random_state=42),oob_score=True,random_state=42)
ens_rf = RandomForestClassifier(random_state=42, **best_param_dict['RandomForest'])
ens_lgbm = lgb.LGBMClassifier(random_state=42, **best_param_dict['LGBM'])
#ens_xbg_model = xgb.XGBClassifier(random_state=42, **best_param_dict['XGB'])
stack = StackingCVClassifier(classifiers=(ens_dt, ens_rf, ens_lgbm),
meta_classifier=ens_rf,random_state=42)
print('5-fold cross validation score:\n')
print('---- {} ----'.format('Stacking Model'))
cv_score = cross_val_score(stack, X, y, cv=5).mean()
print('cv_score :', cv_score)
5-fold cross validation score:
---- Stacking Model ----
cv_score : 0.8485440203329828
[ 부록 ]
Classification Using Neural Network
머신러닝 모델 말고, 딥러닝을 이용하여 분류도 해보았다. 차이점은, input으로 넣을 때 전부 scaling을 해줬다는 점이다. (neuralnet의 경우에는 feature의 scale에 민감하므로)
1. Importing Data
import pandas as pd
import numpy as np
train = pd.read_csv('train.csv')
train.drop('Id', axis=1, inplace=True)
2. Scaling data
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
sc = StandardScaler()
def scalingcolumns(df, cols_to_scale):
for col in cols_to_scale:
df[col] = pd.DataFrame( sc.fit_transform(pd.DataFrame(train[col])),columns=[col] )
return df
cols_to_scale = train.columns[0:-1]
scaled_data = scalingcolumns(train,cols_to_scale)
scaled_data_x = scaled_data.iloc[:,:-1]
scaled_data_y = scaled_data.iloc[:,-1]
scaled_data_y = scaled_data_y-1
- train과 test는 8:2로 나누었다.
x_train,x_test,y_train,y_test = train_test_split(scaled_data_x,scaled_data_y, test_size=0.2,random_state=42)
3. Modeling
import torch
from torch.autograd import Variable
import torchvision.transforms as transforms
import torch.nn.init
import torch.nn as nn
import torch.utils.data as data_utils
# setting hyper parameter
lr = 0.02
training_epochs = 400
batch_size = 288
#keep_prob = 0.7
train.shape
(15120, 55)
trainx = torch.tensor(x_train.values.astype(np.float32))
trainy = torch.tensor(y_train.values)
train_tensor = data_utils.TensorDataset(trainx, trainy)
train_loader = data_utils.DataLoader(dataset = train_tensor, batch_size = batch_size, shuffle = True)
- 다음과 같은 4개의 layer를 쌓았다.
linear1 = nn.Linear(54, 54, bias=True)
linear2 = nn.Linear(54, 54, bias=True)
linear3 = nn.Linear(54, 54, bias=True)
linear4 = nn.Linear(54, 7, bias=True)
- batch normalization
bn1 = nn.BatchNorm1d(54)
bn2 = nn.BatchNorm1d(54)
bn3 = nn.BatchNorm1d(54)
bn4 = nn.BatchNorm1d(7)
relu = nn.ReLU()
# BN으로 이미 regularization 충분!
#dropout = nn.Dropout(p=1 - keep_prob)
- weight의 초기값으로는 He초기값을 사용하였다. ( nn.init.kaiming_uniform )
nn.init.kaiming_uniform_(linear1.weight, mode='fan_in', nonlinearity='relu')
nn.init.kaiming_uniform_(linear2.weight, mode='fan_in', nonlinearity='relu')
nn.init.kaiming_uniform_(linear3.weight, mode='fan_in', nonlinearity='relu')
nn.init.kaiming_uniform_(linear4.weight, mode='fan_in', nonlinearity='relu')
#nn.init.xavier_uniform_(linear1.weight)
#nn.init.xavier_uniform_(linear2.weight)
#nn.init.xavier_uniform_(linear3.weight)
#nn.init.xavier_uniform_(linear4.weight)
- 마지막에는 multi-class classification을 위해 Softmax Function을 사용하였다
model = nn.Sequential(linear1, bn1, relu,
linear2, bn2, relu,
linear3, bn3, relu,
linear4, bn4, nn.Softmax())
- Loss Function으로는 Cross Entropy를, Optimizer로는 Adam을 사용하였다.
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
for epoch in range(training_epochs):
avg_cost = 0
total_batch = x_train.shape[0] // batch_size
for i, (batch_xs, batch_ys) in enumerate(train_loader):
X = Variable(batch_xs.view(288,54))
Y = Variable(batch_ys)
optimizer.zero_grad()
hypothesis = model(X)
cost = criterion(hypothesis, Y.long())
cost.backward()
optimizer.step()
avg_cost += cost/total_batch
if epoch%10==0:
print("[Epoch: {:>4}] cost = {:>.9}".format(epoch, avg_cost.data))
[Epoch: 0] cost = 1.72366369
[Epoch: 10] cost = 1.38898396
[Epoch: 20] cost = 1.35898578
[Epoch: 30] cost = 1.34380758
[Epoch: 40] cost = 1.33171129
[Epoch: 50] cost = 1.32044339
...
[Epoch: 360] cost = 1.24475861
[Epoch: 370] cost = 1.24499512
[Epoch: 380] cost = 1.24003696
[Epoch: 390] cost = 1.2397511
testx = torch.tensor(x_test.values.astype(np.float32))
testy = torch.tensor(y_test.values)
4. Result
(1) train accuracy
trainpred = model(trainx)
correct_trainpred = torch.max(trainpred.data, 1)[1] == trainy.data
correct_trainpred
tensor([1, 1, 1, ..., 1, 1, 1], dtype=torch.uint8)
trainacc = correct_trainpred.float().mean()
print("Accuracy :", trainacc)
Accuracy : tensor(0.9396)
(2) test accuracy
pred = model(testx)
correct_pred = torch.max(pred.data, 1)[1] == testy.data
correct_pred
tensor([1, 0, 1, ..., 1, 1, 1], dtype=torch.uint8)
accuracy = correct_pred.float().mean()
print("Accuracy :", accuracy)
Accuracy : tensor(0.8644)
Overfitting이 발생한 것으로 보인다. 이를 해결하기 위해 drop out도 사용해봤지만, 오히려 성능이 더 좋지 않게 나왔다.
