Semi-unsupervised Learning for TSC
Contents
- Abstract
- Introduction
- Datasets
- HAR (Human Activity Recognition)
- ECG Heartbeat Classification
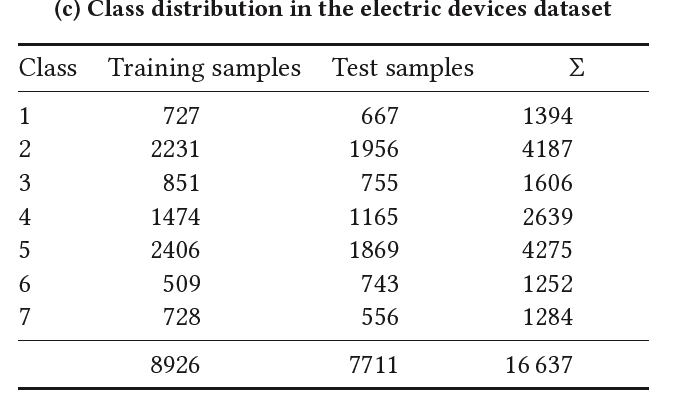
- Electric Devices
- Methodology
- GMM
- GMM for Semi-unsupervised Classification
- Experiments
0. Abstract
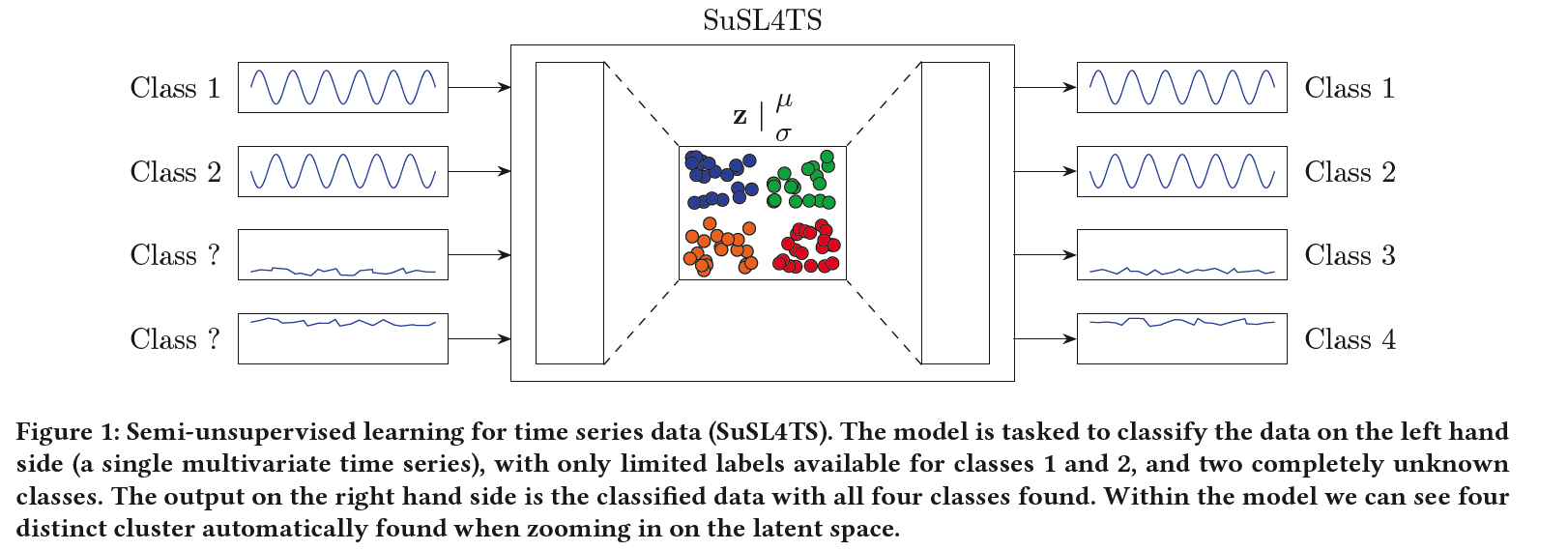
SuSL4TS
-
deep generative GMM for semi-unsupervised learning
-
classify TS data
-
detect sparsely labeled classes (semi-supervised)
& identify emerging classes hidden in the data (unsupervised).
1. Introduction
(1) VAE
- encode the data distribution in the latent space
- allows training on all variations of data
- can see anomaly detection as a probability rather than a raw score
(2) Classification
-
with the development of semi-supervised generative models…able with small amount of data
-
problem : need to know all manifestations of classes beforehand.
(3) Clustering
- could cluster the data, needing no label information at all
- problem : lower classification accuracy & need to manually annotate the found clusters
(4) Classification + Clustering : “hybrid approach of semi-unsupervised learning”
\(\rightarrow\) present SuSL4TS
- a convolutional GMM for semi-unsupervised learning on TS data
Contributions
-
(1) model capable of semi-unsupervised TSC
-
(2) We show the efficacy of our approach on several benchmark datasets
2. Datasets
only hand-select some datasets for our purposes.
3 datasets
- both in the univariate and multivariate setting
- stemming from different domains of data acquisition
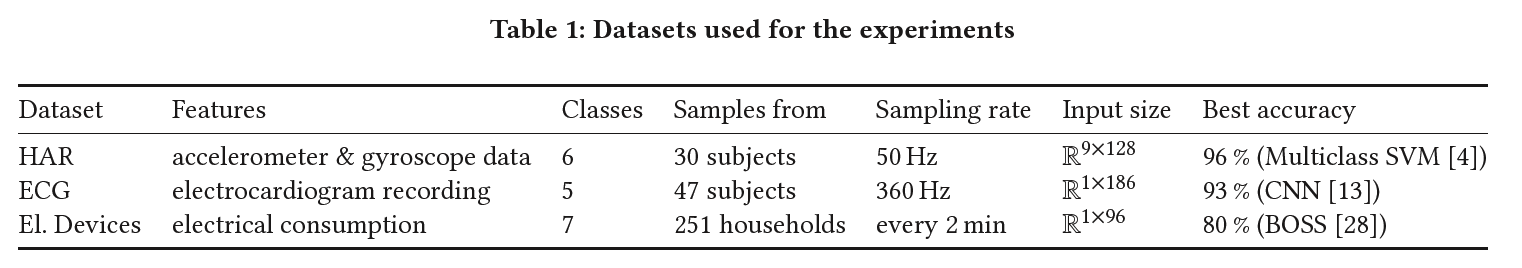
(1) HAR (Human Activity Recognition)
-
consists of data collected from accelerometer and gyroscope sensors in smartphones.
-
# of subjects = 30
-
tasked with performing various Activities of Daily Living (ADL)
-
instructed to perform six distinct ADL adhering to a defined protocol outlining the order of activities.
( standing, sitting, laying down, walking, walking downstairs and walking upstairs )
-
each activity was performed for 15 sec
- except… walking up and downstairs : 12 sec
-
each activity was performed twice & 5 sec pauses separated activities
-
-
pre-processed for noise reduction
( + gravitational and body motion was separated using a low-pass filter )
-
9 signals were sampled with a window of 2.56 sec with 50 % overlap
( i.e. input is of size \(R^{9×128}\) )
-
A feature vector was obtained from each sampling window
- 561 features were extracted
-
70/30 training/test split = 7352 / 2947 samples
(2) ECG Hearbeat Classification
- consists of electrocardiogram (ECG) recordings
-
# of subjects = 47
-
grouped in 5 categories
-
class frequency is skewed towards the \(N\) class ( = Normal )
-
each entry in the set consists of a single heartbeat padded with zeroes
( i.e. input is of size \(R^{1×186}\) )
(3) Electric Devices
-
Samples are taken every 2 min from 251 households
-
After pre-processing and resampling to 15 min averages….
\(\rightarrow\) length of 96 values ( i.e. input is of size \(R^{1×96}\) )
-
regrouping the originally 10 classes to 7
3. Methodology
(1) GMM
- encoder of VAE : \(\Phi(x): \mathbb{R}^n \rightarrow \mathbb{R}^d\)
- decoder of VAE : \(\Theta(z): \mathbb{R}^d \rightarrow \mathbb{R}^n\)
- compressed space is often used for other downstream tasks
- 2 steps
- step 1) unsupervised
- step 2) supervised ( downstream )
-
2-step process can be merged into one
\(\rightarrow\) by adapting the joint probability distribution \(p_{\Theta}\)
\(\rightarrow\) resulting in a Gaussian Mixture Deep Generative model (GMM)
( capable of learning semi-supervised classification )
(2) GMM for Semi-unsupervised Classification
-
adapt [7, 30] and replace 2d conv \(\rightarrow\) 1d conv
- use the work shown in [30] in 2 ways
- 1) use it a reference in performance for the presented convolutional model
- 2) adapt their idea of **Gaussian \(L_2\) reg )
- overall loss function :
- \(\begin{aligned} \mathcal{L} &:=\underset{x, y \in D_l}{\mathbb{E}}\left[\mathcal{L}_l(x, y)-\alpha \cdot \log q_{\Phi}(y \mid x)\right] \\ &+\underset{x \in D_u}{\mathbb{E}}\left[\mathcal{L}_u(x)-\gamma \cdot \lambda \cdot \sum_{c \in C} q_{\Phi}(c \mid x) \cdot \log q_{\Phi}(c \mid x)\right] \\ &+w \cdot \Theta_t . \end{aligned}\).
Notation
- \(D_l\) : labeled subset of the data
- \(D_u\) : all unlabeled data
- \(\Theta_t\) : trainable weights at epoch \(t\)
- \(\alpha, \gamma, \lambda\) : hyperparameters weighting the entropy regularization
- loss terms \(\mathcal{L}_l, \mathcal{L}_u\) measure the evidence lower bound (ELBO) from the GMM model
