[ 1. Introduction : ML for Graphs ]
( 참고 : CS224W: Machine Learning with Graphs )
Contents
-
1-1. Why Graphs
-
1-2. Applications of Graph ML
-
1-3. Choice of Graph Representation
1-1. Why Graphs
1) Graphs
= general language for describing & analyzing entities with…
- 1) “relations”
- 2) “interactions”
Lots of data are “graphs”!
- ex) computer network, event graphs,…
2) Types of networks & graphs
Networks ( = Natural Graphs )
- 1) Social networks ( society )
- 2) Communication & Transactions
- 3) Biomedicine ( ex. genes )
- 4) Brain connections ( ex. thoughts )
Graphs ( = representation )
- 1) information/knowledge
- 2) similarity networks
- 3) relational structures
Relational Graphs
-
model better relationships between entities!
-
use DL for complex modeling
3) Networks are complex
Why is it complex?
- arbitrary size & complex structure
- no fixed node ordering or reference point
- dynamic & multi-modal features
4) DL in graphs
[ Input ]
- Network
[ Output ] (Prediction)
- 1) node labels
- 2) new links
- 3) generated graphs / subgraphs
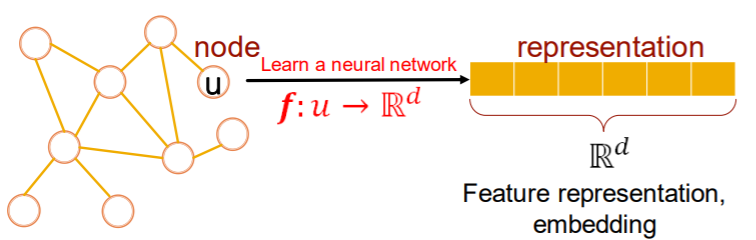
Key point : “Representation Learning”
-
“automatically” learn features with DNN
-
map to \(d\)-dim embeddings
- similar nodes = similar in embedded space
5) Outline
- 1) Traditional methods
- Graphlets, Graph Kernels
- 2) Node Embeddings
- DeepWalk, Node2vec
- 3) GNN
- GCN, GraphSAGE, GAT
- 4) Knowledge graph & reasoning
- TransE, BetaE
-
5) Deep Generative models ( for graphs )
- 6) Applications
1-2. Applications of Graph ML
1) Diverse Tasks
- 1) Node classification
- ex) categorizing users/items
- 2) Link prediction
- ex) knowledge graph completion
- 3) Graph Classification
- ex) Molecule property prediction
- 4) Clustering
- ex) community detection
- 5) etc
- Graph generation, Graph evolution ..
Diverse level of tasks
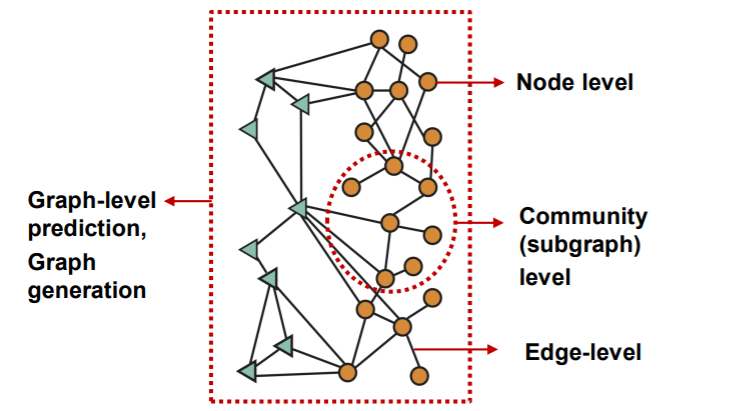
2) Example of “NODE-level” ML
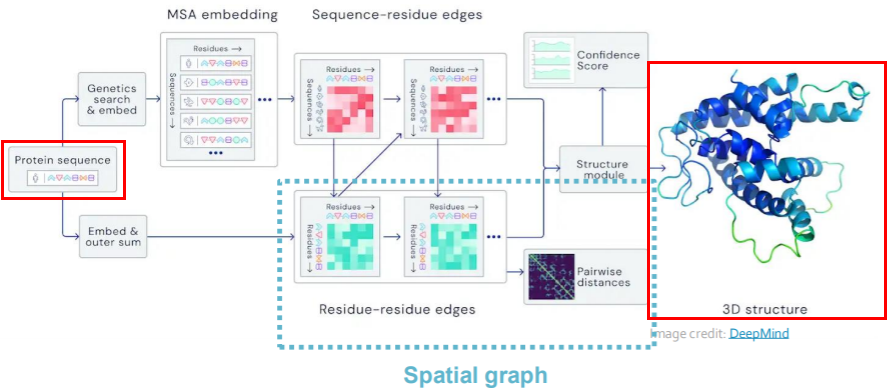
ex) Protein Folding
- Protein
- protein= sequence of amino acid
- 3d structure
- interact with each other
- Goal : “predict 3D structure”, based on “amino acid sequence”
- key idea of AlphaFold : “spatial graph”
- (1) node : amino acids
- (2) edges : proximity between nodes
3) Example of “EDGE-level” ML
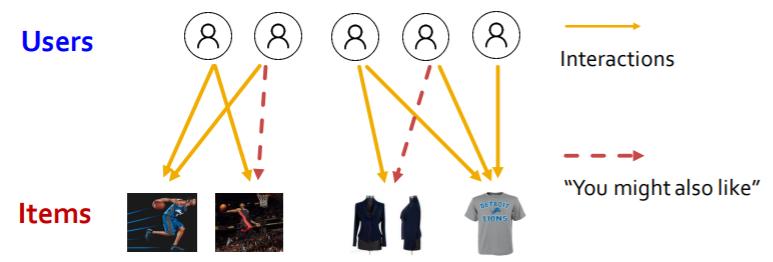
ex) Recommender Systems
- Formulation
- (1) node : user & items
- (2) edge : user & item interaction
-
Goal : “Recommend item to users”
( predict whether 2 nodes are related )
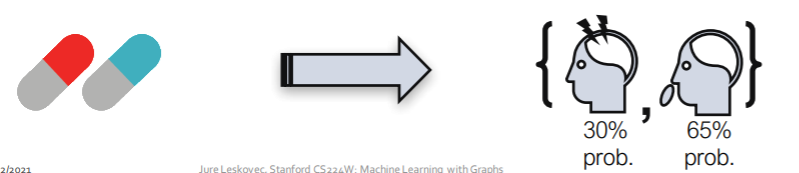
ex) Drug Side Effects
- Background : many patients & many drugs
-
Goal : predict adverse side effects of “pair of drugs”
- Formulation
- (1) node : drugs & proteins
- (2) edges : interactions
- drug-protein interaction
- protein-protein interaction
- drug-drug interaction
4) Example of “SUBGRAPH-level” ML
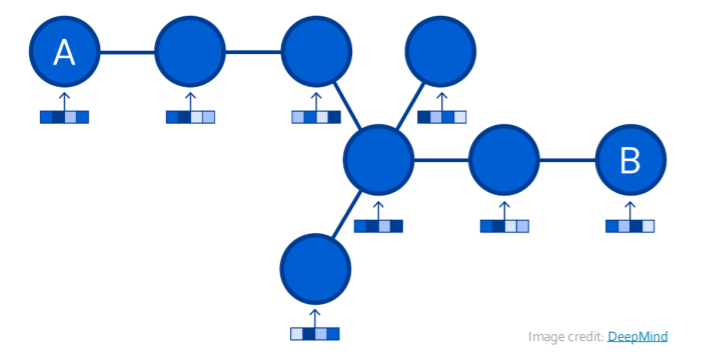
ex) Traffic Prediction
- want to move from A to B…. how long will it take?
- Formulation
- (1) node : road segments
- (2) edges : connectivity between nodes
- ex) Google Maps : traffic prediction with GNN
5) Examples of “GRAPH-level” ML
ex) Drug Discovery
- Antibiotics = small molecular graphs
- Formulation
- (1) node : atoms
- (2) edges : chemical bonds
- (Q) Which molecules should be prioritized?
- ex) graph classification model
- predict promising molecules among candidates
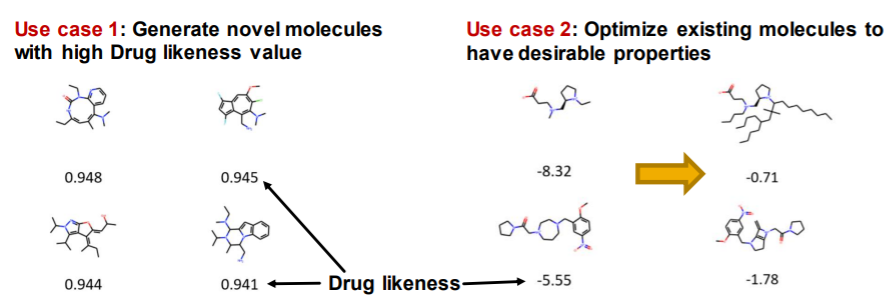
ex) Graph generation
- generate novel molecules ( new structure )
- with “high drug likeness”
- with “desirable properties”
ex) Physics Simulation
- Formulation
- (1) node : particles
- (2) edges : interaction between nodes
- Goal : predict how a graph will EVOLVE
1-3. Choice of Graph Representation
1) Components of Network
-
(1) Objects : nodes, vertices ( notation : \(N\) )
-
(2) Interactions : links, edges ( notation : \(E\) )
-
(3) System : network, graph ( notation : \(G(N,E)\) )
Notation :
- \(\mid N \mid\) : number of nodes
- \(\mid E \mid\) : number of edges
[ Question ]
Which one is proper representation??
How to build a graph?
- 1) what are nodes?
- 2) what are edges?
[ Answer ]
- differ by domain/tasks!
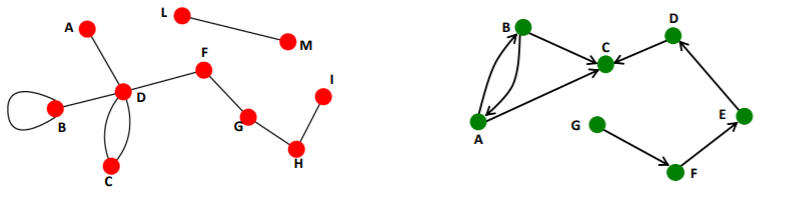
2) Directed & Undirected Graphs
Directed
- links = arcs
- ex) phone calls, following in INSTAGRAM
Undirected
- links = symmetrical, reciprocal
- ex) collaborations, friendship in FB
3) Node Degree
Node degree ( \(k_i\) )
- # of edges, adjacent to node \(i\)
Average degree
- \(\bar{k}=\langle k\rangle=\frac{1}{N} \sum_{i=1}^{N} k_{i}=\frac{2 E}{N}\).
Directed Network
In-degree & Out-degree
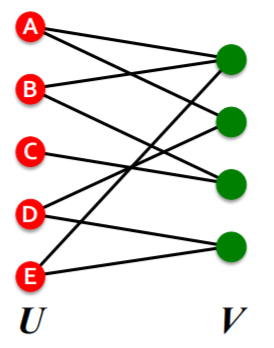
4) Bipartite Graph
graph, whose nodes can be divided into “2 disjoint sets \(U\), \(V\)”, where …
every link connects a node in \(U\) to one in \(V\)
( = \(U\) & \(V\) : independent sets )
ex) User-Movie rating
Folded / Projected Bipartite Graphs
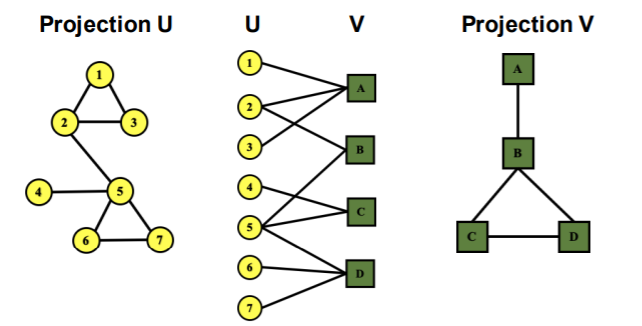
5) Adjacency Matrix
Different ways of representing graphs :
-
1) adjacency matrix
-
2) edge list & adjacency list
Adjacency Matrix
\(n \times n\) matrix ( \(n\) : number of nodes )
- connected = 1
- disconnected = 0
( Undirected Graph : symmetric adjacency matrix )
Problem : Adjacency matrices are SPARSE!
6) Edge list & Adjacency list
Different ways of representing graphs :
-
1) adjacency matrix
-
2) edge list & adjacency list
(a) Edge list
- ex) (2,3), (2,4), (3,2), (3,4) ,… (5,2)
(b) Adjacency list
- easier when network is large & sparse
- ex)
- 1 :
- 2 : 3,4
- 3 : 2,4
- 4 : 5
- 5 : 1,2
7) Node & Edge ATTRIBUTES
ex) weight, ranking, type, sign, …
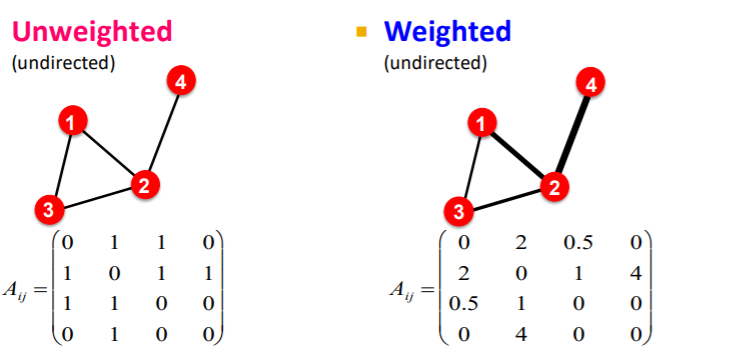
8) More Types of Graphs
unweighted & weighted
- unweighted ( 1 or 0 )
- weighted ( not binary )
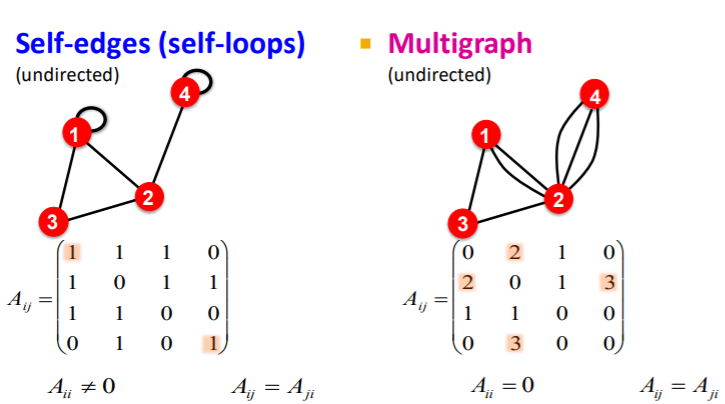
self-edges & multi-graph
- self-edges ( self-loops )
- multi-graph
- multiple edges between pair of nodes
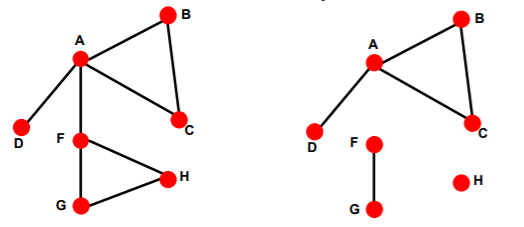
connected & disconnected graph
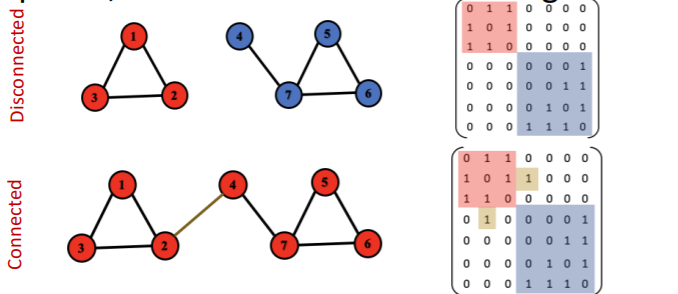
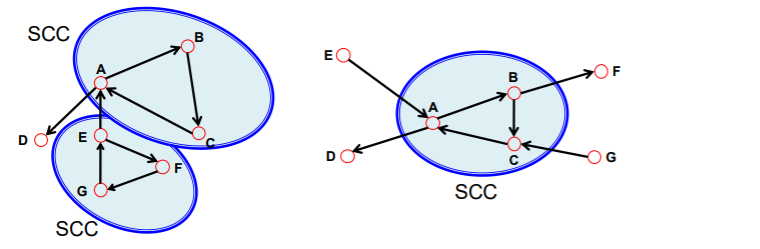
strongly & weakly connected graph
- strongly : both A-B & B-A path
- weakly : disregard edge direction
Strongly connected components (SCCs)