[ 14. Traditional Generative Models for Graphs ]
( 참고 : CS224W: Machine Learning with Graphs )
Contents
- Properties of Real-world Graphs
- Erdos-Renyi Random Graphs
- The Small-world Model
- Kronecker Graph Model
- Summary
Graph Generative models
Why is it needed?
- Insights : understand ‘formulation of graph’
- Predictions : predict how the ‘graph will evolve’
- Simulations
- Anomaly detection
Road map of Graph Generation
[1] Properties of Real-world Graphs
- properties for successful graph generative model
[2] Traditional graph generative models
- assumption on graph formulation process
- ex) Erdos-Renyi Random Graphs
[3] Deep graph generative models
- (in lecture 15)
1. Properties of Real-world Graphs
(1) Key Network properties
- Degree distribution : \(P(k)\)
- Clustering coefficient : \(C\)
- Connected components : \(s\)
- Path length : \(h\)
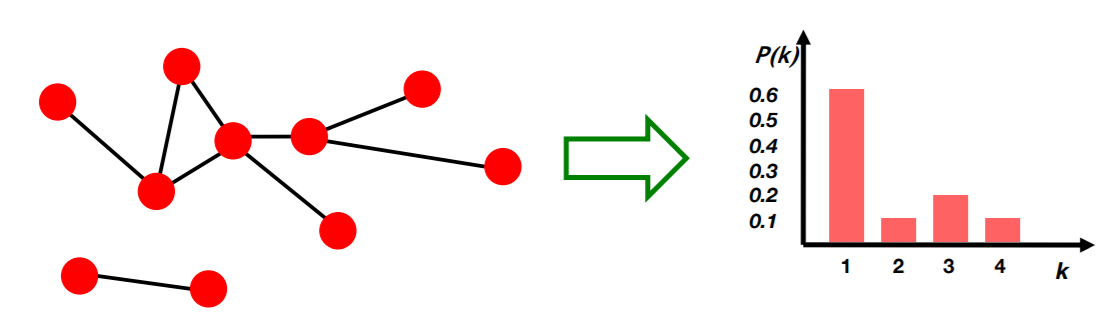
a) Degree distribution : \(P(k)\)
probability that a randomly chosen node has degree k
- Notation :
- \(N_k\) : # of nodes with “degree k”
- \(P(k)\) : normalized version ( = \(N_k / N\) )
b) Clustering coefficient : \(C\)
how are node \(i\)’s’ neighbors connected to each other?
-
Notation
- \(k_i\) : degree of node \(i\)
- \(C_i\) : clustering coefficient of node \(i\) ( = \(\frac{2e_i}{k_i(k_i-1)}\) )
- \(e_i\) = # of edges between the “node \(i\)’s’ neighbors”
-
Example :
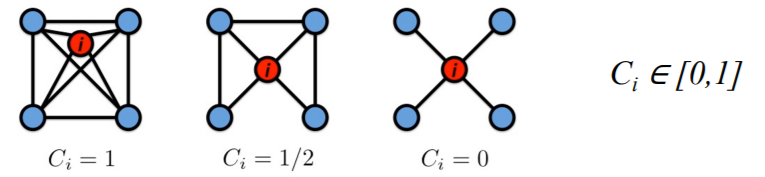
Graph Clustering Coefficient?
- take average of all “node clustering coefficient” ( \(C = \frac{1}{N}\sum_{i=1}^{N}C_i\) )
c) Connected components : \(s\)
Size of the “LARGEST connected component”
- Connected component : set where any two nodes can be joined by a path
- How to find “connected component”?
- step 1) select random node
- step 2) BFS (Breadth-First Search)
- label all the visited nodes
- visited nodes are connected
- step 3) if unvisited nodes exists,
- repeat step 1) & step 2)
d) Path length : \(h\)
Diameter = maximum (shortest path) distance between “ANY” pair of nodes
- Notation
- \(h_{ij}\) : distance from node \(i\) & \(j\)
- \(E_{max}\) : max # of possible edges ( = \(n(n-1)/2\) )
- \(\bar{h}\) : Average path length
- Average path length :
- \(\bar{h}=\frac{1}{2 E_{\max }} \sum_{i, j \neq i} h_{i j}\).
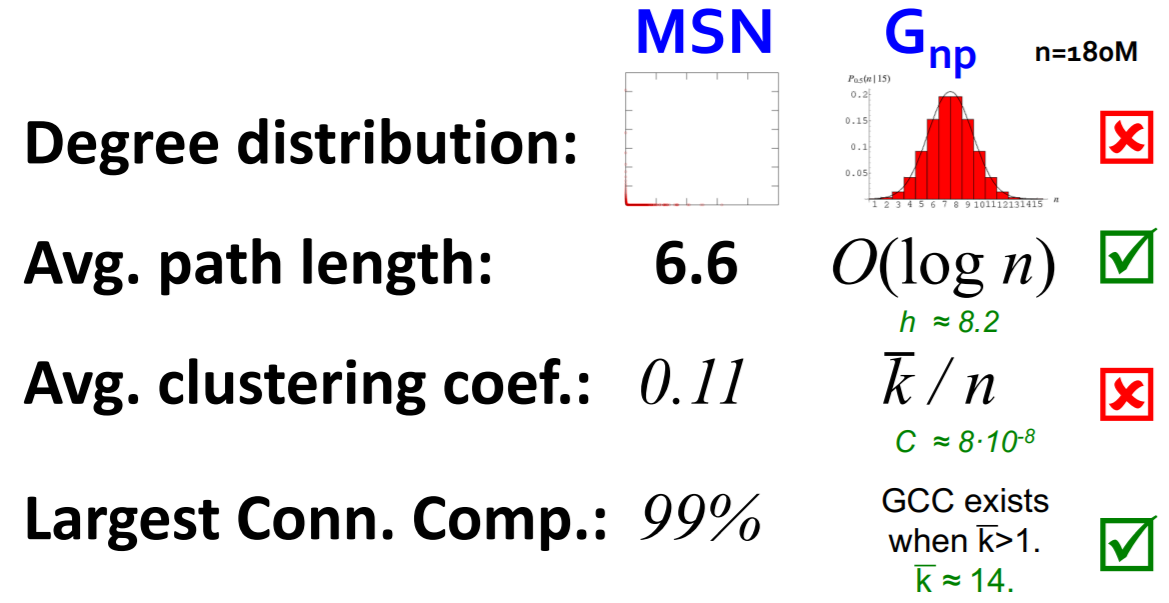
(2) Case Study : MSN graph
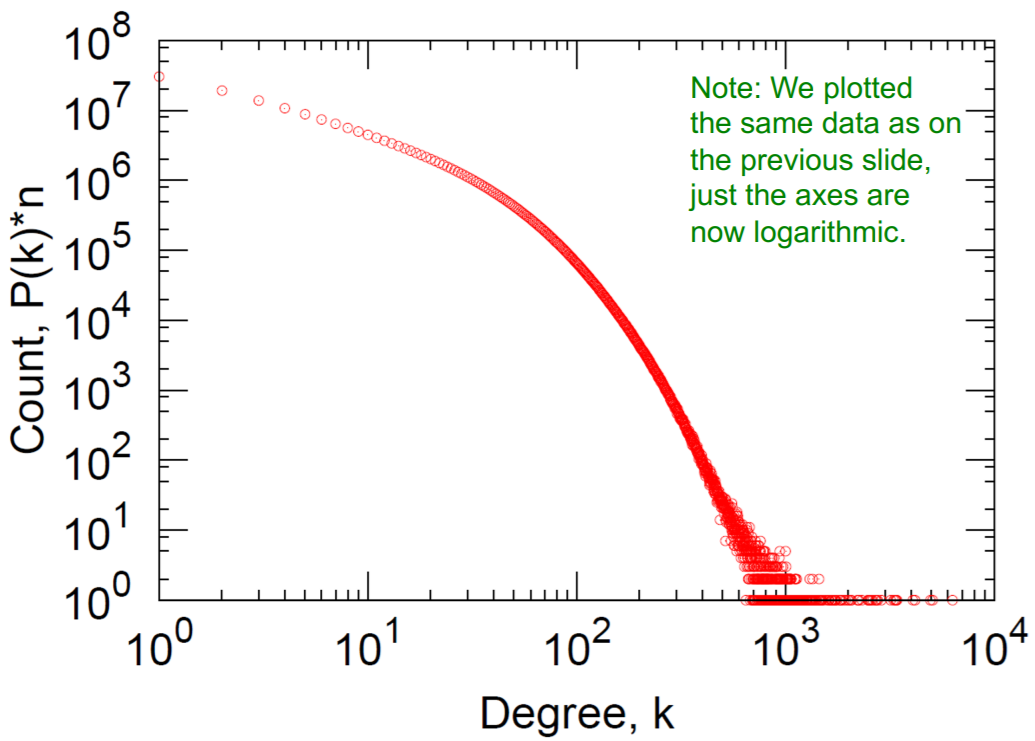
a) Degree distribution
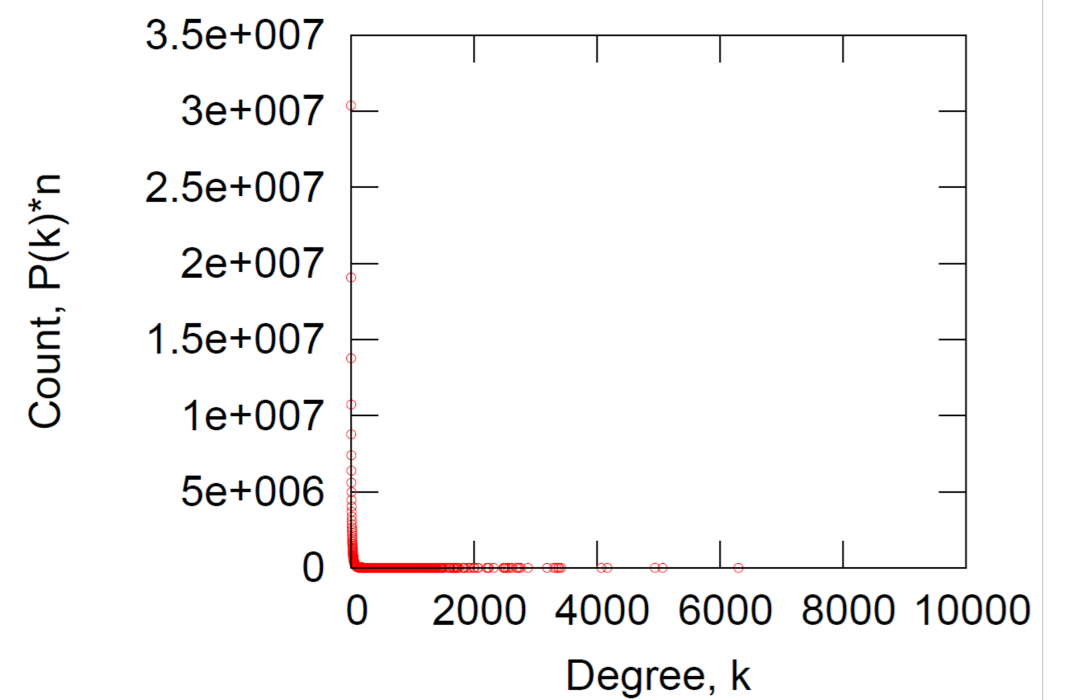
Log-Log Degree distribution
b) Clustering coefficient
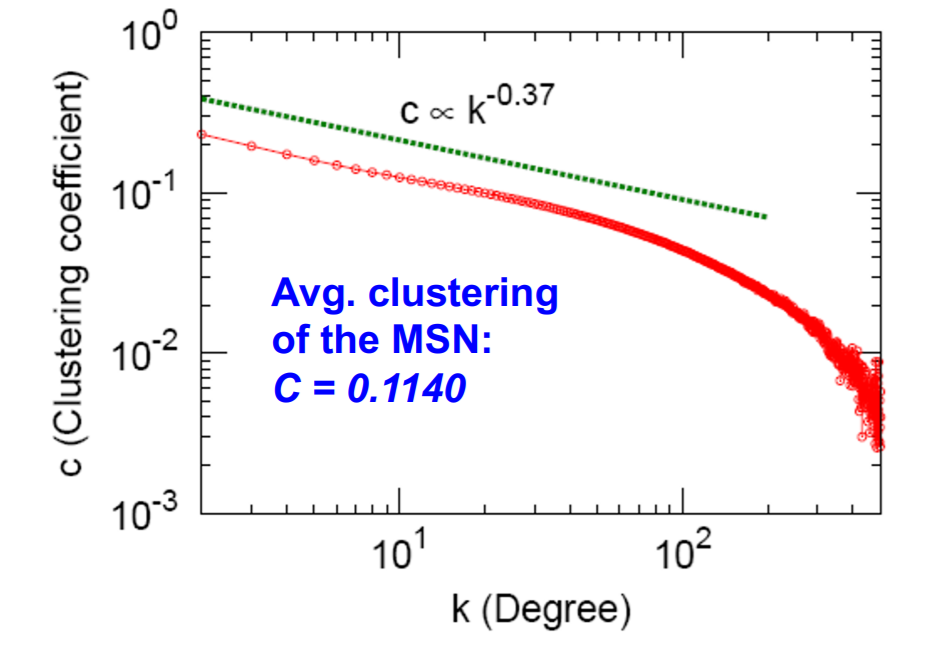
\(C_k\) : “average \(C_i\), of nodes \(i\) with degree k”
- \(C_k = \frac{1}{N}\sum_{i:k_i=k}^{}C_i\).
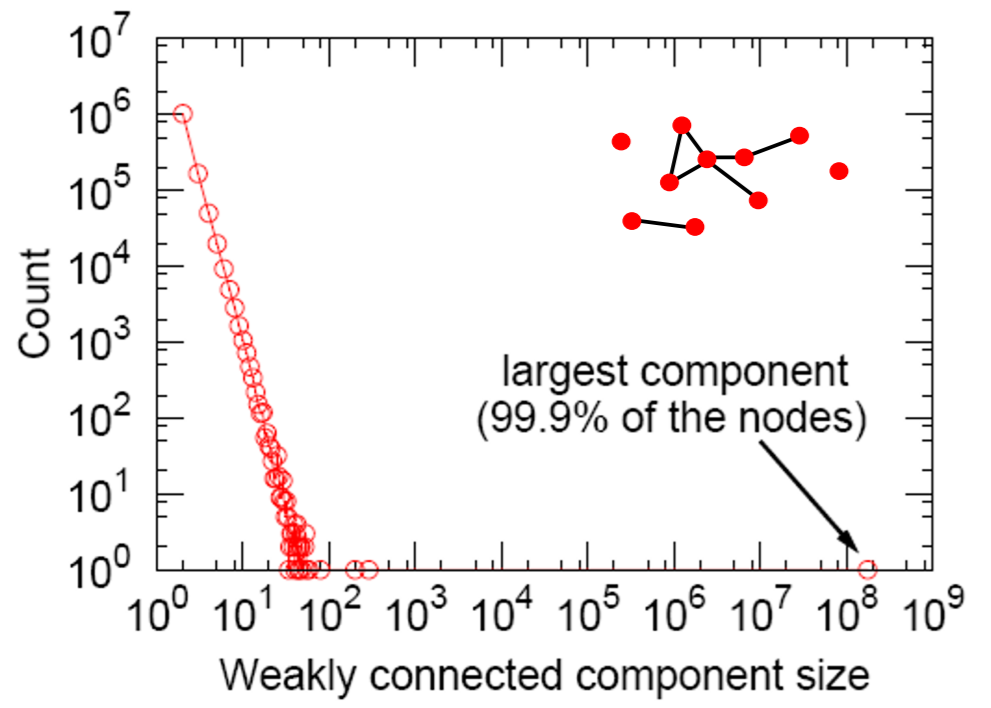
c) Connected Components
d) Path Length
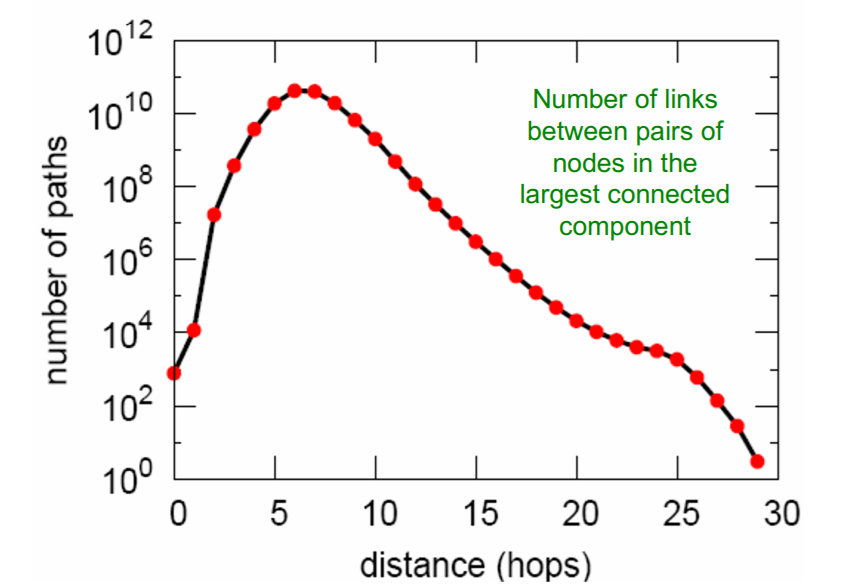
-
Average Path Length = 6.6
-
90% of nodes can be reached in \(< 8\) hops
Summary
- a) Degree distribution : heavily skewed (avg degree=14.4)
- b) Clustering coefficient : 0.11
- c) Connectivity : giant component (99%)
- d) Path length : 6.6
Question : so, is it a surprising/expected result?
\(\rightarrow\) need a NULL model
2. Erdos-Renyi Random Graphs
(1) Introduction
-
Simplest model of graphs
-
Two variants
- 1) \(G_{np}\) : \(n\) nodes & prob \(p\) of connected
- 2) \(G_{nm}\) : \(n\) nodes & \(m\) edges
\(\rightarrow\) expected edges are same, but 1) is stochastic, 2) is deterministic
( = 1) is “RANDOM” graph model / not unique )
(2) Random Graph model \(G_{np}\)
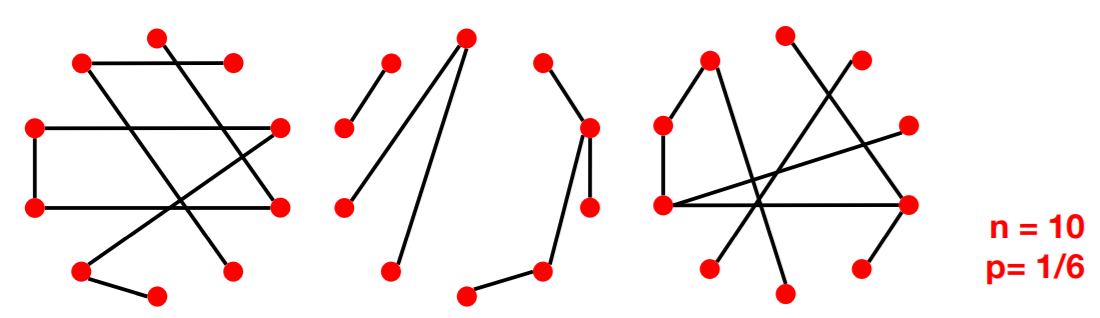
Properties of \(G_{np}\)
- 1) Degree distribution : \(P(k)\)
- 2) Clustering coefficient : \(C\)
- 3) Connected Component : \(s\)
- 4) Path length : \(h\)
(3) Degree distribution : \(P(k)\)
Binomial Distribution : \(k \sim \text{Binom}(n-1,p)\)
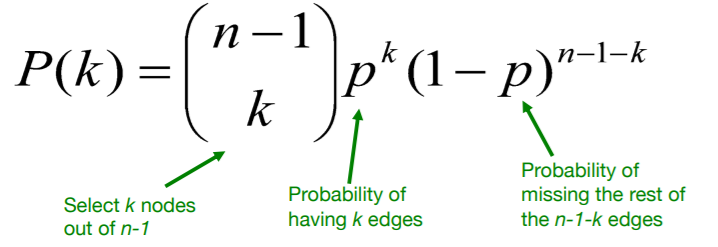
- mean : \(\bar{k}=p(n-1)\).
- variance : \(\sigma^{2}=p(1-p)(n-1)\)
(4) Clustering coefficient : \(C\)
Clustering coefficient : \(C_{i}=\frac{2 e_{i}}{k_{i}\left(k_{i}-1\right)}\)
- \(k_i\) : degree of node \(i\)
Expected \(\boldsymbol{e}_i\) : \(\mathrm{E}\left[\boldsymbol{e}_{\boldsymbol{i}}\right] =p \frac{k_{i}\left(k_{i}-1\right)}{2}\)
- 1) \(p\) : prob of connection
- 2) \(\frac{k_{i}\left(k_{i}-1\right)}{2}\) : # of distinct pairs of neighbors of node \(i\) of degree \(k_i\)
- 1) clustering coefficient of random graph is SMALL
- 2) fixed average degree + bigger graph \(\rightarrow\) decrease of \(C\)
(5) Connected Component of \(G_{np}\)
How does “change of \(p\)” influence the “graph structure”?
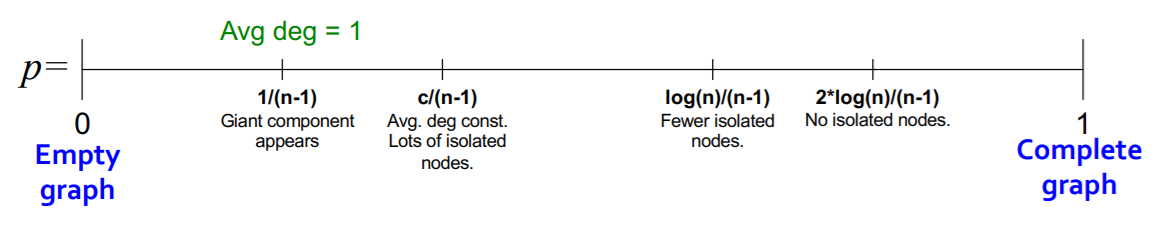
Giant component occurs ( = GCC exists ) , when…
\(\rightarrow\) average degree \(k\) is bigger than 1
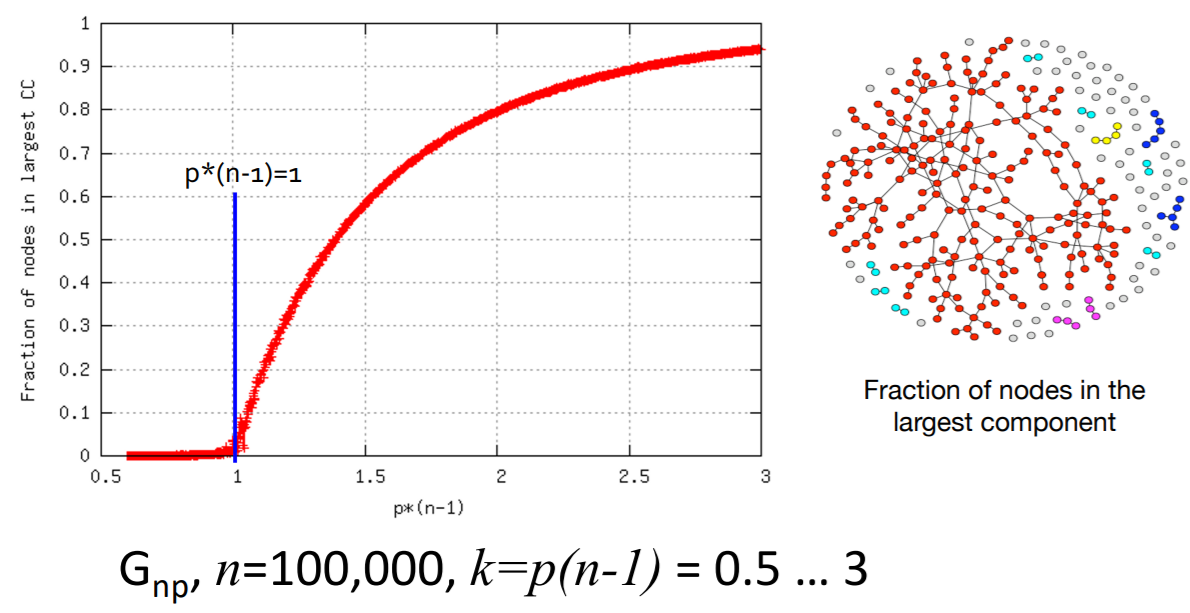
(6) Path length : \(h\)
need to know the notion of “EXPANSION”
Expansion
-
graph \(G(V,E)\) has expansion \(\alpha\) =
if \(\forall S \subseteq V\), # of edges leaving \(S \geq \alpha \cdot \min (\mid S \mid, \mid V \backslash S \mid)\)
-
that is…
\[\alpha=\min _{S \subseteq V} \frac{\text { #edges leaving } S}{\min ( \mid S \mid , \mid V \backslash S \mid )}\]
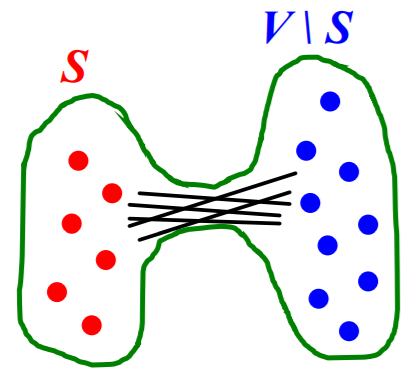
Meaning :
- measure of “robustness”
- to disconnect \(l\) nodes, we need to cut \(\geq \alpha \cdot l\) edges
Example :
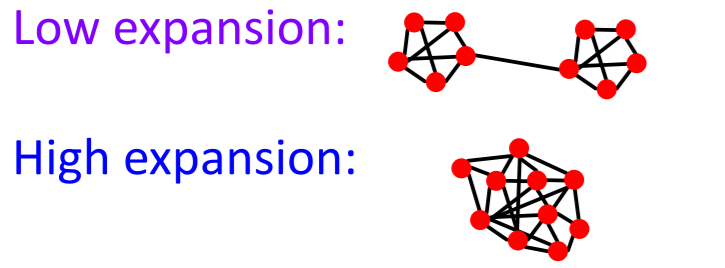
Details
-
graph with \(n\) nodes & \(\alpha\) expansion
= for all pairs of nodes, “there is a path of length \(O((log n)/\alpha)\)“
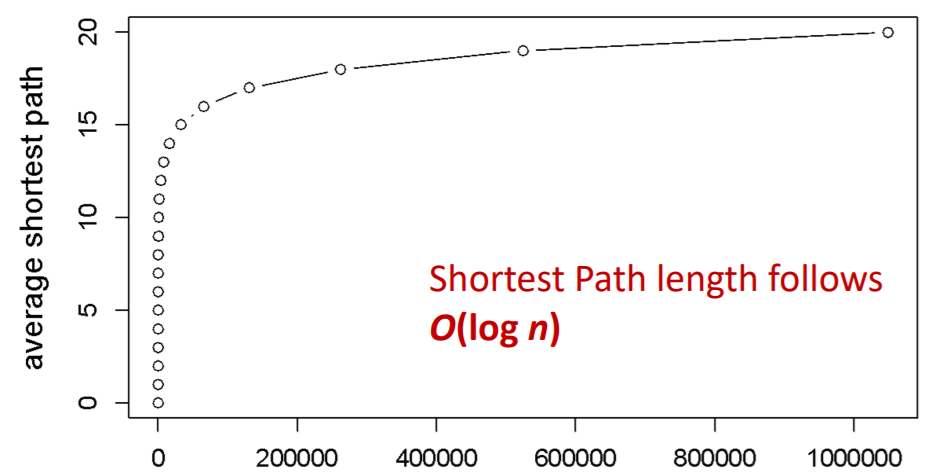
Shortest path of \(G_{np}\)
- ER random graph can grow very large, but nodes will be just a few hops apart
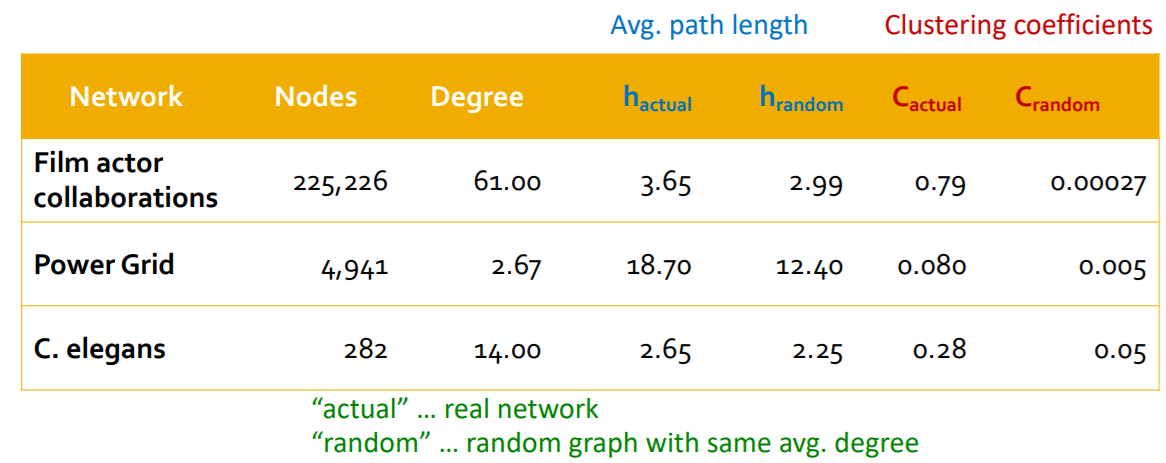
(7) MSN vs \(G_{np}\)
3. The Small-world Model
(1) Motivation for small-world
Trade-off of “clustering coefficient” & “diameter”?
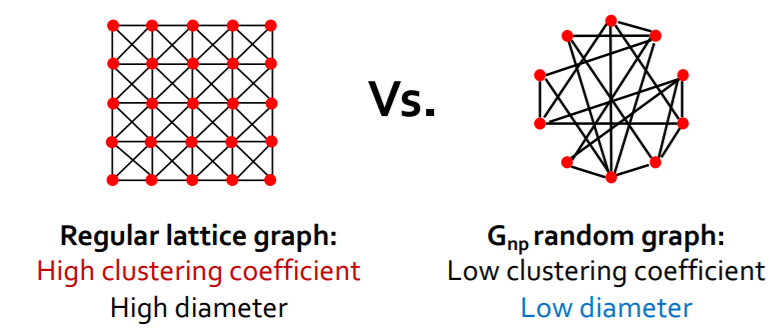
\(\rightarrow\) can we have “HIGH clustering” & “LOW(SHORT) diameter” ?
Real networks have “HIGH clustering”
- ex) MSN have \(7\) times higher \(C\) than \(G_{np}\)
Examples :
(2) Interpolate “regular lattice” & “\(G_{np}\)”
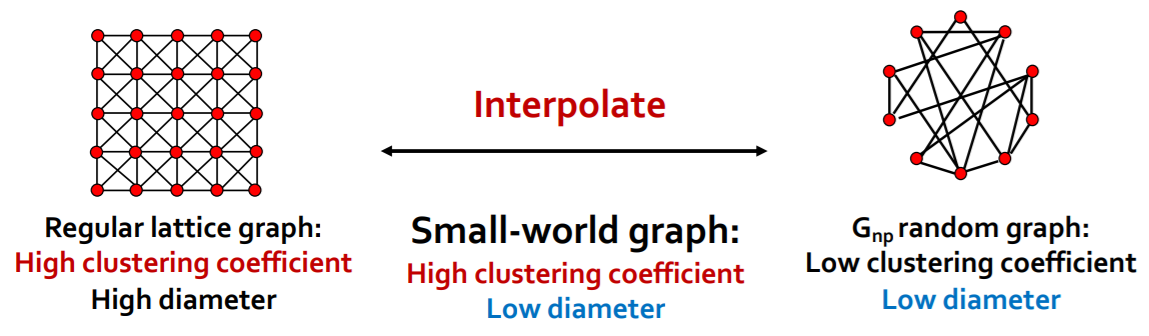
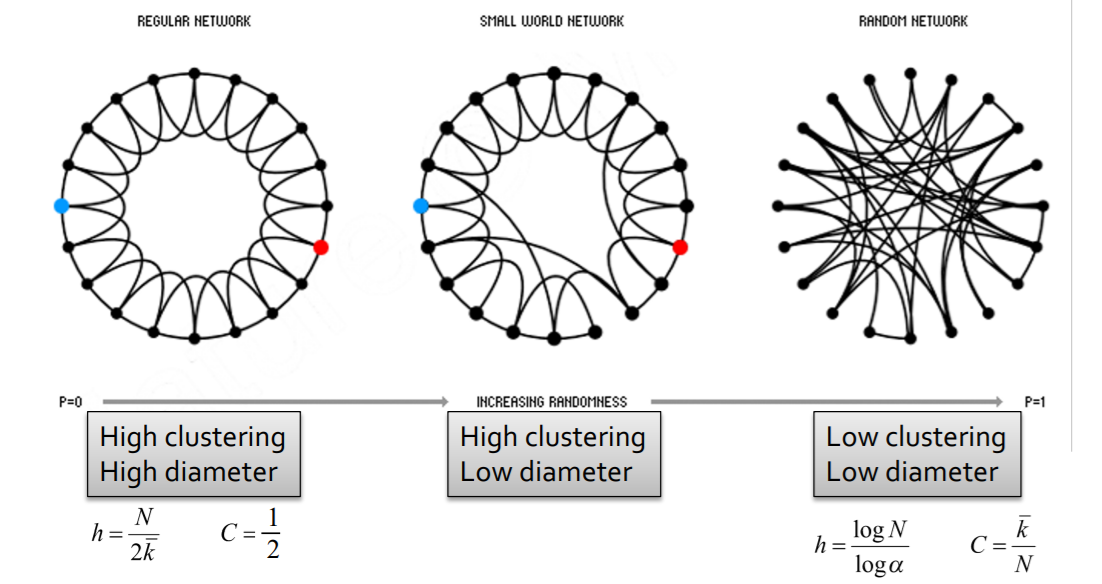
Solution : “Small-world Model”
[Step 1] start with low-dim regular lattice
- high clustering coefficient
[Step 2] REWIRE
-
introduce randomness (shortcuts)
-
add/remove edges ( to join remote parts )
( with prob \(p\) )
-
-
enables “interpolation between 2 types of graphs”
4. Kronecker Graph Model
Think of network structure recursively! ( key : self-similarity )
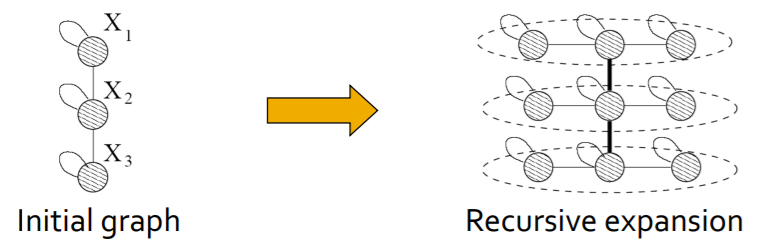
\(\rightarrow\) by using “Kronecker product”
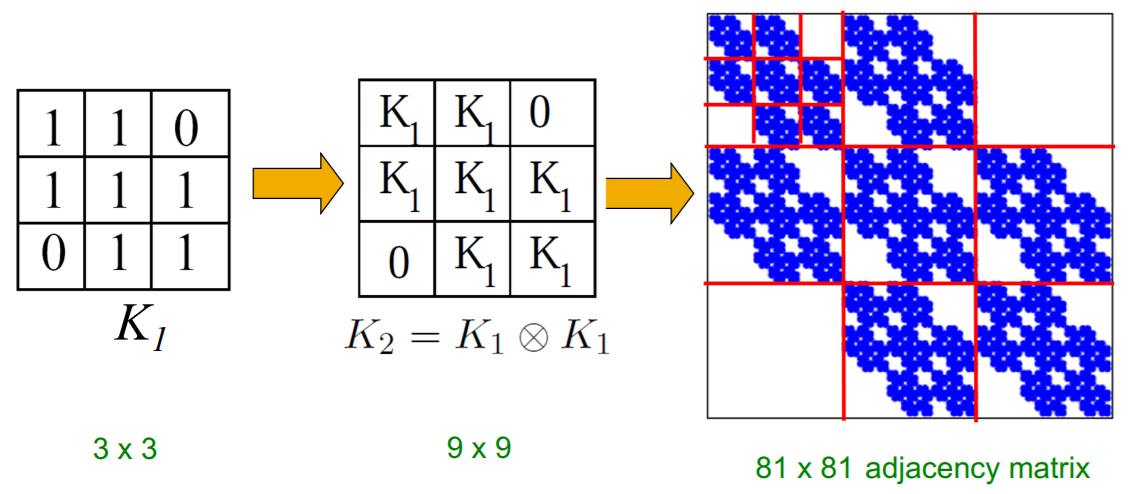
(1) Kronecker Product
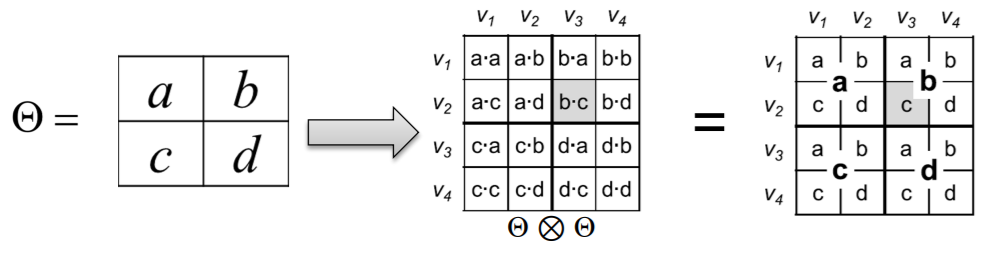
\(\mathbf{C}=\mathbf{A} \otimes \mathbf{B} \doteq\left(\begin{array}{cccc} a_{1,1} \mathbf{B} & a_{1,2} \mathbf{B} & \ldots & a_{1, m} \mathbf{B} \\ a_{2,1} \mathbf{B} & a_{2,2} \mathbf{B} & \ldots & a_{2, m} \mathbf{B} \\ \vdots & \vdots & \ddots & \vdots \\ a_{n, 1} \mathbf{B} & a_{n, 2} \mathbf{B} & \ldots & a_{n, m} \mathbf{B} \end{array}\right)\).
- Kronecker product of two graphs
= Kronecker product of their adjacency matrices
- \(K_{1}^{[\mathrm{m}]}=K_{\mathrm{m}}=\underbrace{K_{1} \otimes K_{1} \otimes \ldots K_{1}}_{\mathrm{m} \text { times }}=K_{\mathrm{m}-1} \otimes K_{1}\).
(2) Kronecker Graph
“recursive” model of network structure
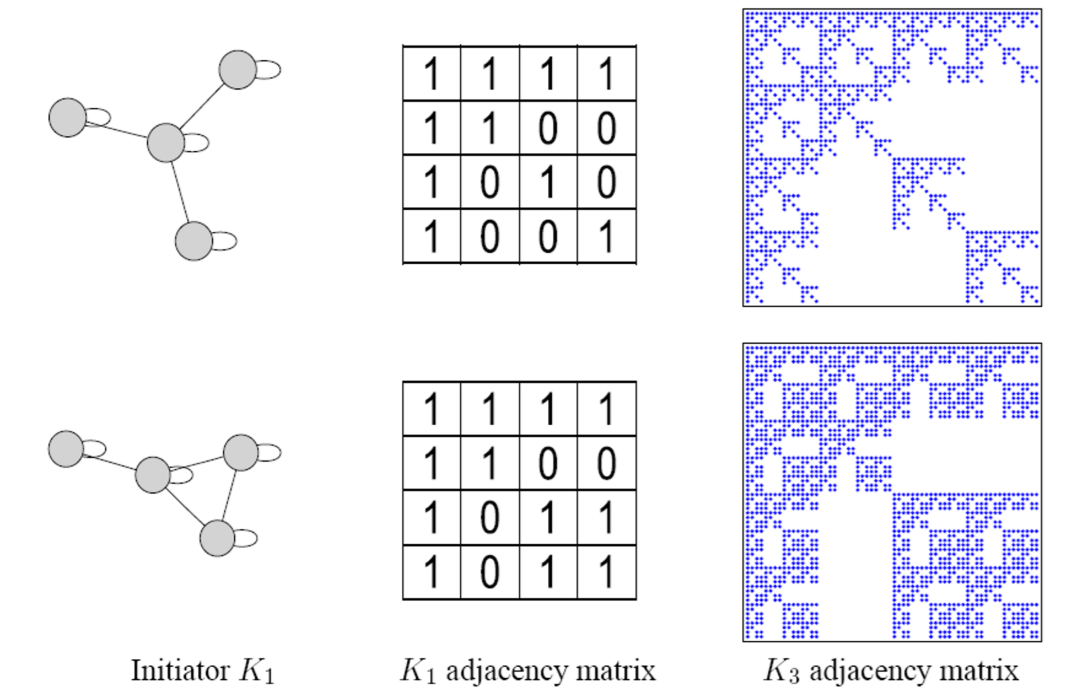
(3) Stochastic Kronecker Graph
[Step 1] create \(N_1 \times N_1\) probability matrix \(\Theta_1\)
[Step 2] compute \(k^{th}\) Kronecker power \(\Theta_k\)
[Step 3] include edge \((u,v)\) , with prob \(p_{uv}\)
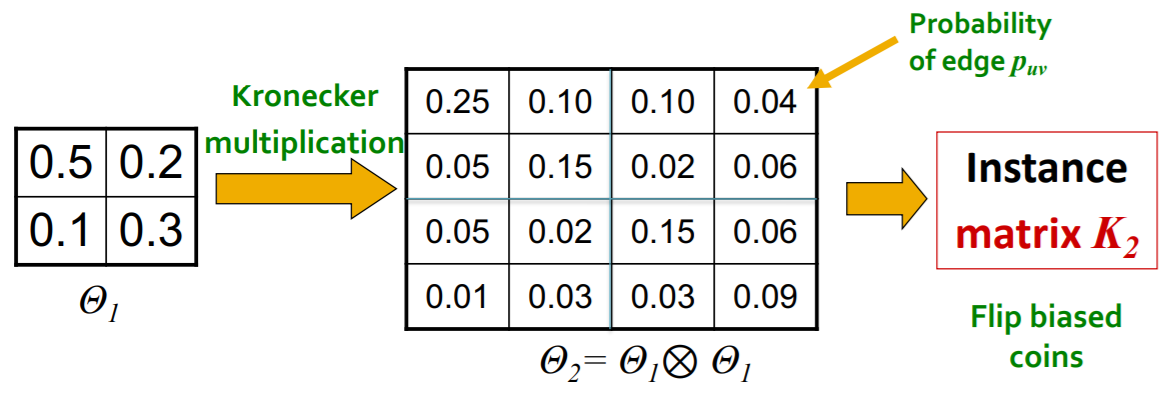
(4) Generation of Kronecker Graphs
Just as “coin flip”!
- exploit the recursive structure of Kronecker graph
5. Summary
Traditional graph generative models
- 1) Erdös-Renyi graphss
- 2) Small-world graphs
- 3) Kronecker graphs
\(\rightarrow\) they all have an assumption ofgraph generation processes
