GC-LSTM : GCN embedded LSTM for Dynamic Network Link Prediction (2018)
Contents
- Abstract
- Methodology
- Problem Definition
- Overall Framework
- GC-LSTM model
- Decoder Model
- Loss Function
0. Abstract
Dynamic Network Link Prediction
- challenge : network structure evolves with time
Propose GCN embedded LSTM ( = GC-LSTM ) for dynamic link prediction
- LSTM : for temporal features of all snapshots of dynamic network
-
each snapshot : GCN is applied to capture local structural properties
- can predict both ADDED & REMOVED links
- \(\leftrightarrow\) existing methods : only handle removed links
1. Methodology
(1) Problem Definition
Dynamic Networks
sequence of discrete snapshots : \(\left\{G_{1}, \cdots, G_{T}\right\}\)
- \(G_{t}=\left(V, E_{t}, A_{t}\right)(t \in[1, T])\)……. network at time \(t\)
- \(V\) : nodes
- \(E_{t}\) : temporal links within the fixed timespan \([t-\tau, t]\)
Static Network
-
aims to predict the future links by the current network
-
focus on structural feature of the network
\(\leftrightarrow\) Dynamic network : also need to learn the temporal feature of the network
Goal
-
extract the structural feature of each snapshot network through GCN
-
learn temporal structure via LSTm
Network Link Prediction in Dynamic Networks
Dynamic network link prediction
-
structural sequence modeling problem
-
aims to learn the evolution information of the previous \(T\) snapshots,
to predict the probability of all links at time t
-
\(\hat{A}_{t}=\operatorname{argmax} P\left(A_{t} \mid A_{t-T}, \cdots, A_{t-1}\right)\).
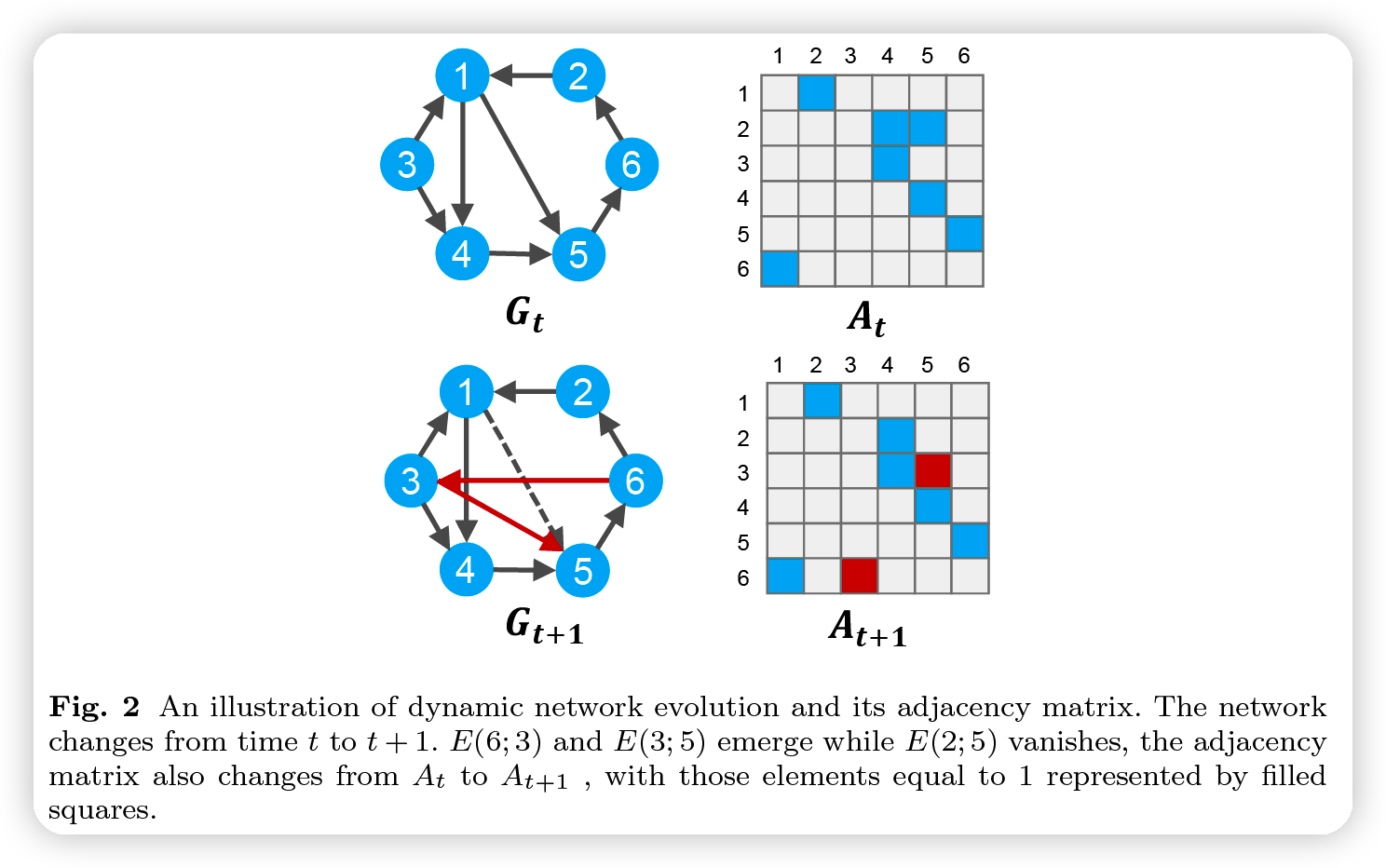
(2) Overall Framework
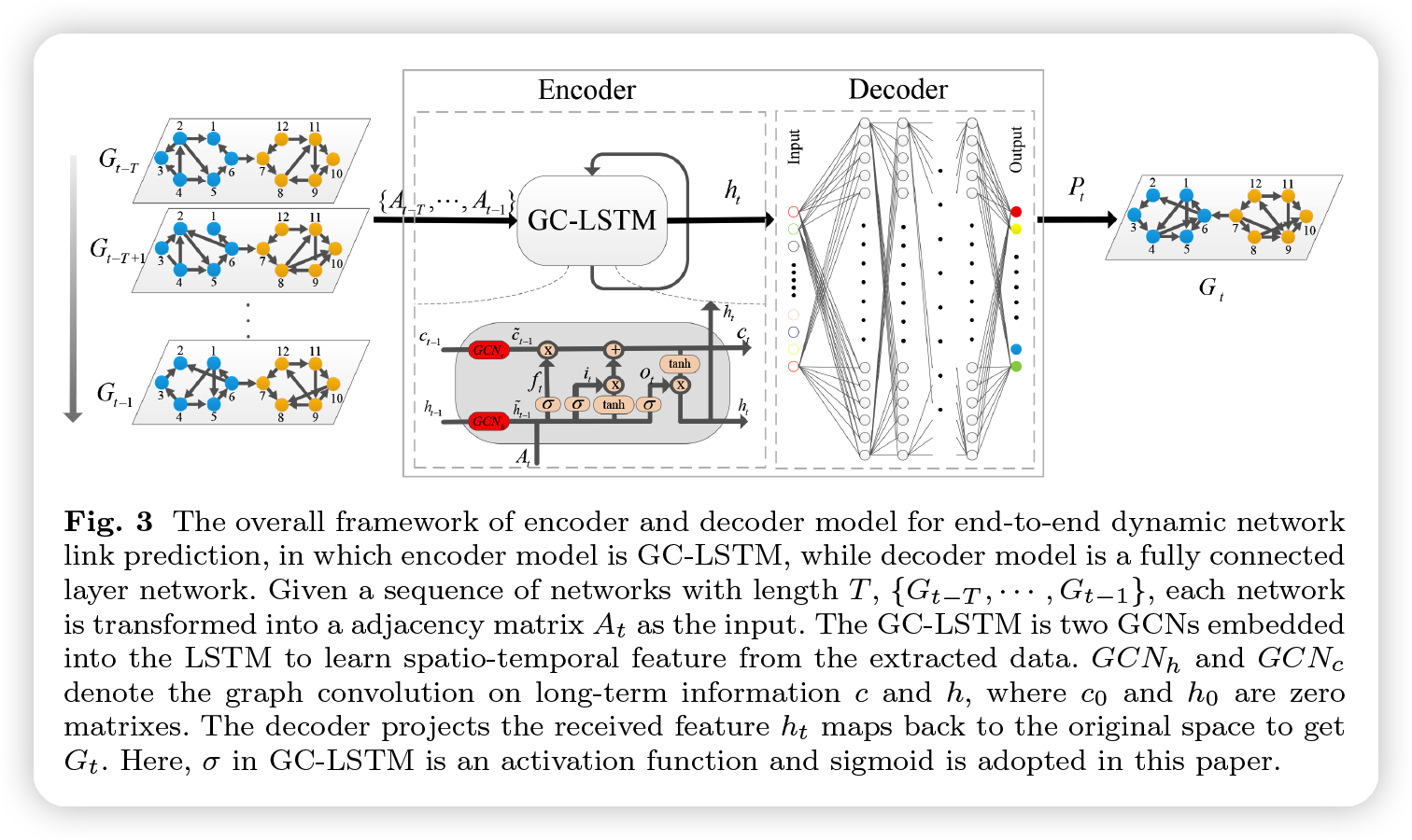
GC-LSTM
- encoder & decoder
- encoder = GCN embedded LSTM
- GCN : learn network structure
- LSTM : learn temporal information
- decoder = FC layer
- convert feature map to original space
(3) GC-LSTM model
Input
- input sequence data : \(\left\{A_{t-T}, \cdots, A_{t-1}\right\}\)
- last hidden layer vector : \(h_{t}\)
Dynamic Network Link prediction
-
linkage status of each node with others at multiple times
= regarded as TIME SERIES
ChebNet : \(g_{\theta}=\sum_{k=0}^{K} \theta_{k} T_{k}(\tilde{L})\)
GC-LSTM model
-
mainly relies on 2 state values
- (1) hidden state \(h\)
- cell state \(c\)
-
in the dynamic network link prediction task,
need to consider the influence of \(h\) of neighbors on \(h\) of target
& influence of \(c\) of negibhros
\(\rightarrow\) thus, propose to use 2 GCN models, for \(h\) & \(c\)
Steps
[ step 1 ] Decide what info will be thrown away frm previous cell state
- by forget gate …… \(f_{t} \epsilon[0,1]^{d}\)
- \(f_{t}=\sigma\left(A_{t} W_{f}+G C N_{f}^{K}\left(\tilde{A}_{t-1}, h_{t-1}\right)+b_{f}\right)\).
[ Step 2 ] Update the cell state
- (1) \(\bar{c}_{t} \epsilon[-1,1]^{d}\) : tanh layer generates a new candidate vector of the cell layers,
- (2) \(i_{t} \epsilon[0,1]^{d}\) : sigmoid layer which determines how many new candidate vector will be added to the cell state
- (3) \(c_t\) : update cell state ( by (1) & (2) )
\(\begin{aligned} \bar{c}_{t} &=\tanh \left(A_{t} W_{c}+G C N_{o}^{K}\left(\tilde{A}_{t-1}, h_{t-1}\right)+b_{c}\right) \\ i_{t} &=\sigma\left(A_{t} W_{i}+G C N_{c}^{K}\left(\tilde{A}_{t-1}, h_{t-1}\right)+b_{i}\right) \\ c_{t} &=f_{t} \odot G C N_{c}^{K} c_{t-1}+i_{t} \cdot \bar{c}_{t} \end{aligned}\).
[ Step 3 ] Decide output ( by output gate )
\(\begin{aligned} &o_{t}=\sigma\left(A_{t} W_{o}+G C N_{o}^{K}\left(\tilde{A}_{t-1}, h_{t-1}\right)+b_{0}\right) \\ &h_{t}=o_{t} \odot \tanh \left(c_{t}\right) \end{aligned}\).
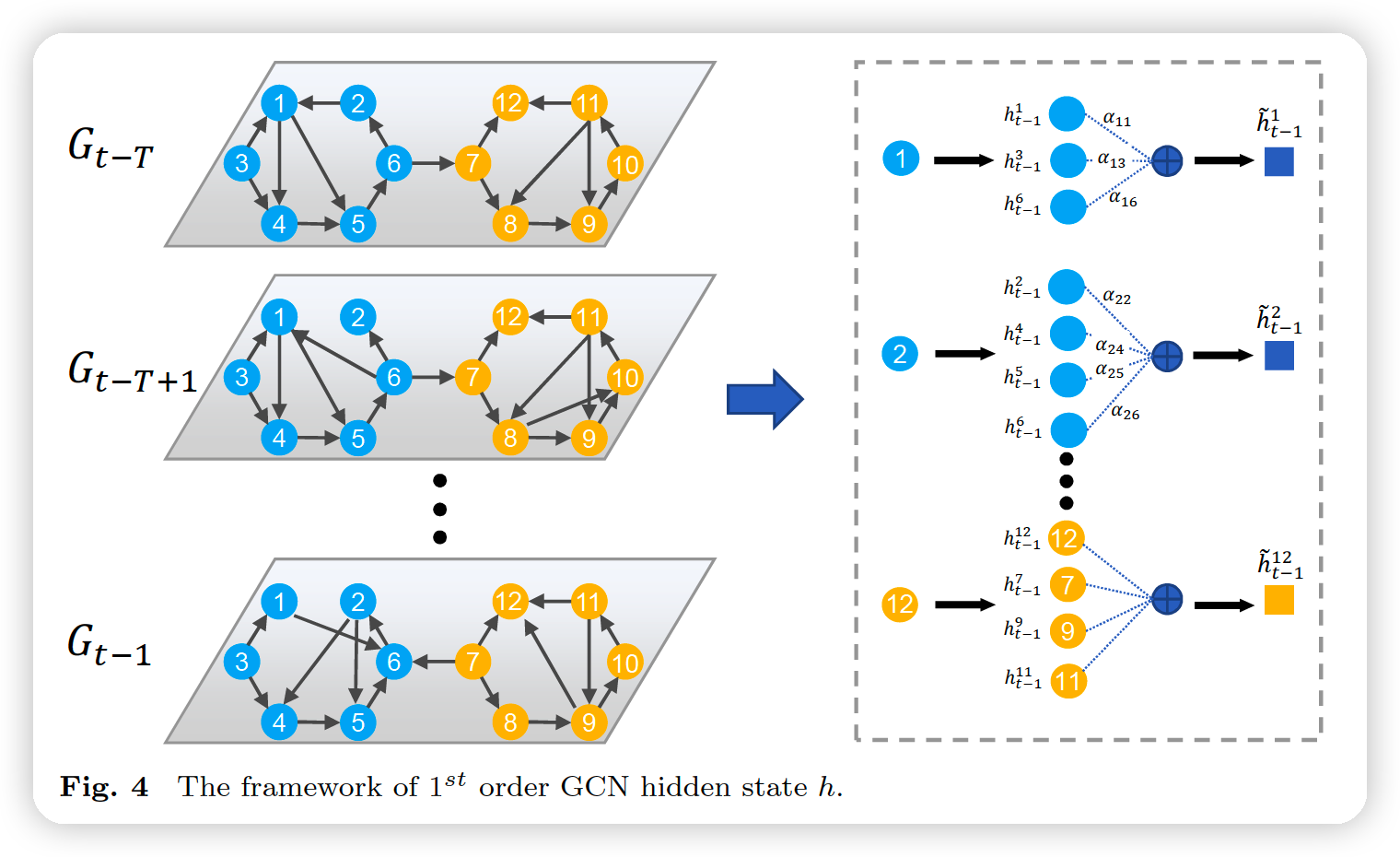
(4) Decoder model
\(\begin{aligned} &Y_{d}^{(1)}=\operatorname{ReLU}\left(W_{d}^{(1)} h_{t}+b_{d}^{(1)}\right) \\ &Y_{d}^{(k)}=\operatorname{ReLU}\left(W_{d}^{(k)} Y_{d}^{(k-1)}+b_{d}^{(k)}\right) \\ &P_{t}=o_{t} \odot \tanh \left(c_{t}\right) \end{aligned}\).
(5) Loss Function
\(L_{2}= \mid \mid P_{t}-A_{t} \mid \mid_{F}^{2}=\sum_{i=0}^{N} \sum_{j=0}^{N}\left(P_{t}(i, j)-A_{t}\right)\).
\(L=L_{2}+\beta L_{r e g}\).
