( Skip the basic parts + not important contents )
12.Continuous Latent Variables
Ch 9) Probabilistic models with “DISCRETE” latent variables
Ch 12) Probabilistic models with “CONTINUOUS” latent variables
12-1. PCA (Principal Component Analysis)
PCA : used in..
- dimension reduction
- lossy data compression
- feature extraction
- data vizualization
2 common definitions of PCA
-
1) orthogonal projection of data into lower dimension linear space ( = principal subspace )
-
2) linear projection that minimizes the average projection cost
( = mean squared distance between data points & their projections)
12-1-1. Maximum variance formulation
\(D\) : original dimension
\(M\) : reduced dimension
example with 1D reduction (\(M=1\)):
- \(u_1\) : \(D\) dim vector
- \[u_1^Tu_1=1.\]
projected data
- mean : \(\mathbf{u}_{1}^{\mathrm{T}} \overline{\mathbf{x}}\)
- where \(\overline{\mathbf{x}}=\frac{1}{N} \sum_{n=1}^{N} \mathbf{x}_{n}\)
- variance : \(\frac{1}{N} \sum_{n=1}^{N}\left\{\mathbf{u}_{1}^{\mathrm{T}} \mathbf{x}_{n}-\mathbf{u}_{1}^{\mathrm{T}} \overline{\mathbf{x}}\right\}^{2}=\mathbf{u}_{1}^{\mathrm{T}} \mathbf{S} \mathbf{u}_{1}\)
- where \(\mathbf{S}=\frac{1}{N} \sum_{n=1}^{N}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)^{\mathrm{T}}\)
maximize \(\mathbf{u}_{1}^{\mathrm{T}} \mathbf{S} \mathbf{u}_{1}\) ,w.r.t \(\mathbf{u}_{1}\)
-
with constraint \(\mathbf{u}_{1}^{\mathrm{T}} \mathbf{u}_{1}=1\)
( use Lagrange multiplier )
-
\(\mathbf{u}_{1}^{\mathrm{T}} \mathbf{S} \mathbf{u}_{1}+\lambda_{1}\left(1-\mathbf{u}_{1}^{\mathrm{T}} \mathbf{u}_{1}\right)\).
\(\mathbf{S u}_{1}=\lambda_{1} \mathbf{u}_{1}\).
\(\therefore \mathbf{u}_{1}^{\mathrm{T}} \mathbf{S} \mathbf{u}_{1}=\lambda_{1}\).
-
Variance will be maximum, if we set \(u_1\) equal to the eigenvector, that has largest eigenvalue \(\lambda_1\)
Generalization to \(M\) dimension
-
\(M\) eigenvectors \(\mathbf{u}_{1}, \ldots, \mathbf{u}_{M}\)
\(M\) largest eigenvalues \(\lambda_{1}, \ldots, \lambda_{M}\)
-
Summary : PCA involves…
- 1) evaluating \(\bar{\mathbf{x}}\) and \(S\) ( covariance matrix )
- 2) find \(M\) eigenvectors of \(S\), corresponding to the \(M\) largest eigenvalues
-
Computational cost
-
full eigenvector decomposition : \(O(D^3)\)
-
if project our data onto \(M\) PCs :
( using efficient techniques, such as “power method” : \(O(MD^2)\) )
( can also use EM algorithm )
-
12-1-2. Minimum-error formulation
alternative formulation of PCA, based on “PROJECTION ERROR MINIMIZATION”
-
introduce complete orthonormal set of \(D\)-dimensional basis vectors \(\{u_i\}\)
( where \(i=1, \ldots, D\) and \(\mathbf{u}_{i}^{\mathrm{T}} \mathbf{u}_{j}=\delta_{i j}\))
each data point can be represented with linear combination
-
\(\mathbf{x}_{n}=\sum_{i=1}^{D} \alpha_{n i} \mathbf{u}_{i}\).
\(\alpha_{n j}=\mathbf{x}_{n}^{\mathrm{T}} \mathbf{u}_{j},\) (by orthonormality property)
\(\therefore\) \(\mathbf{x}_{n}=\sum_{i=1}^{D}\left(\mathbf{x}_{n}^{\mathrm{T}} \mathbf{u}_{i}\right) \mathbf{u}_{i}\).
wish to use representation in restricted number \(M < D\)
approximate each datapoint as below : \(\tilde{\mathbf{x}}_{n}=\sum_{i=1}^{M} z_{n i} \mathbf{u}_{i}+\sum_{i=M+1}^{D} b_{i} \mathbf{u}_{i}\).
- \(\left\{z_{n i}\right\}\) depend on the particular data point
- \(\left\{b_{i}\right\}\) are constants
Goal : minimize \(J=\frac{1}{N} \sum_{n=1}^{N}\left\|\mathbf{x}_{n}-\tilde{\mathbf{x}}_{n}\right\|^{2}\).
-
(1) w.r.t \(z_{ni}\) : \(z_{n j}=\mathbf{x}_{n}^{\mathrm{T}} \mathbf{u}_{j}\), where \(j=1, \ldots, M\)
-
(2) w.r.t \(b_i\) : \(b_{j}=\overline{\mathbf{x}}^{\mathrm{T}} \mathbf{u}_{j}\) , where \(j=M+1, \ldots, D\)
\(\rightarrow\) substitute using (1), (2),
then \(\mathbf{x}_{n}-\tilde{\mathbf{x}}_{n}=\sum_{i=M+1}^{D}\left\{\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)^{\mathrm{T}} \mathbf{u}_{i}\right\} \mathbf{u}_{i}\)
Thus, \(J=\frac{1}{N} \sum_{n=1}^{N}\left\|\mathbf{x}_{n}-\tilde{\mathbf{x}}_{n}\right\|^{2}=\frac{1}{N} \sum_{n=1}^{N} \sum_{i=M+1}^{D}\left(\mathbf{x}_{n}^{\mathrm{T}} \mathbf{u}_{i}-\overline{\mathbf{x}}^{\mathrm{T}} \mathbf{u}_{i}\right)^{2}=\sum_{i=M+1}^{D} \mathbf{u}_{i}^{\mathrm{T}} \mathbf{S} \mathbf{u}_{i}\)
Example with \(D=2\) and \(M=1\)
-
minimize \(J=\mathbf{u}_{2}^{\mathrm{T}} \mathrm{Su}_{2},\)
subject to \(\mathrm{u}_{2}^{\mathrm{T}} \mathbf{u}_{2}=1\)
-
Loss function : \(\tilde{J}=\mathbf{u}_{2}^{\mathrm{T}} \mathbf{S} \mathbf{u}_{2}+\lambda_{2}\left(1-\mathbf{u}_{2}^{\mathrm{T}} \mathbf{u}_{2}\right)\)
\(\rightarrow\) \(\mathbf{S u}_{2}=\lambda_{2} \mathbf{u}_{2}\)
-
Summary
minimizing average squared projection distance = maximum variance
general solution to minimizing \(J\) : \(\mathbf{S u}_{i}=\lambda_{i} \mathbf{u}_{i}\)
distortion measure : \(J=\sum_{i=M+1}^{D} \lambda_{i}\)
-
( = select e.v that those having \(D-M\) smallest eigenvalues )
-
( = choosing principal subspace, corresponding to \(M\) largest eigenvalues)
12-1-3. Applications of PCA
(1) PCA for data compression
PCA approximation to \(\tilde{x_n}\):
\(\begin{aligned} \tilde{\mathbf{x}}_{n} &=\sum_{i=1}^{M}\left(\mathbf{x}_{n}^{\mathrm{T}} \mathbf{u}_{i}\right) \mathbf{u}_{i}+\sum_{i=M+1}^{D}\left(\overline{\mathbf{x}}^{\mathrm{T}} \mathbf{u}_{i}\right) \mathbf{u}_{i} \\ &=\overline{\mathbf{x}}+\sum_{i=1}^{M}\left(\mathbf{x}_{n}^{\mathrm{T}} \mathbf{u}_{i}-\overline{\mathbf{x}}^{\mathrm{T}} \mathbf{u}_{i}\right) \mathbf{u}_{i} \end{aligned}\).
for each datapoint, we have replaced
- before) \(D\) dimension vector \(x_n\)
- after) \(M\) dimension vector with components \(\left(\mathbf{x}_{n}^{\mathrm{T}} \mathbf{u}_{i}-\overline{\mathbf{x}}^{\mathrm{T}} \mathbf{u}_{i}\right)\)
(2) PCA for data pre-processing
-
dimension reduction (X)
standardize (O)
Covariance matrix for standardized data :
- \(\rho_{i j}=\frac{1}{N} \sum_{n=1}^{N} \frac{\left(x_{n i}-\bar{x}_{i}\right)}{\sigma_{i}} \frac{\left(x_{n j}-\bar{x}_{j}\right)}{\sigma_{j}}\).
By using PCA, we can make more substantial normalization,
so that “different variables become decorrelated”
(1) write \(\mathbf{S u}_{i}=\lambda_{i} \mathbf{u}_{i}\) in the form \(\mathbf{S}\mathbf{U}=\mathbf{U}\mathbf{L}\)
- \(\mathbf{L}\) : \(D \times D\) diagonal matrix, with elements \(\lambda_i\)
- \(\mathbf{U}\) : \(D \times D\) orthogonal matrix, with columns \(\mathbf{u}_i\)
(2) transformed value \(\mathbf{y_n}\) : \(\mathbf{y}_{n}=\mathbf{L}^{-1 / 2} \mathbf{U}^{\mathrm{T}}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\)
(3) \(\mathbf{y}_{n}=\mathbf{L}^{-1 / 2} \mathbf{U}^{\mathrm{T}}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\)’s mean and variance
-
mean : 0
-
variance : \(I\) (identity matrix)
\(\begin{aligned} \frac{1}{N} \sum_{n=1}^{N} \mathbf{y}_{n} \mathbf{y}_{n}^{\mathrm{T}} &=\frac{1}{N} \sum_{n=1}^{N} \mathbf{L}^{-1 / 2} \mathbf{U}^{\mathrm{T}}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)^{\mathrm{T}} \mathbf{U} \mathbf{L}^{-1 / 2} \\ &=\mathbf{L}^{-1 / 2} \mathbf{U}^{\mathrm{T}} \mathbf{S} \mathbf{U} \mathbf{L}^{-1 / 2}=\mathbf{L}^{-1 / 2} \mathbf{L} \mathbf{L}^{-1 / 2}=\mathbf{I} \end{aligned}\).
12-1-4. PCA for high-dimensional data
case of \(N<<D\)
\(\rightarrow\) computationally infeasible ( = \(O(D^3)\) )
How to solve?
(1) let \(\mathbf{X}\) to be \(N \times D\) dim, centered matrix
- \(n^{\text{th}}\) row : \(\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)^{\mathrm{T}}\)
(2) Then, \(\mathbf{S}=\frac{1}{N} \sum_{n=1}^{N}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)^{\mathrm{T}}\) can be written as
- \(\mathbf{S}=N^{-1} \mathbf{X}^{\mathrm{T}} \mathbf{X}\).
(3) eigenvector equation
-
\(\frac{1}{N} \mathbf{X}^{\mathrm{T}} \mathbf{X} \mathbf{u}_{i}=\lambda_{i} \mathbf{u}_{i}\).
-
\(\frac{1}{N} \mathbf{X} \mathbf{X}^{\mathrm{T}}\left(\mathbf{X} \mathbf{u}_{i}\right)=\lambda_{i}\left(\mathbf{X} \mathbf{u}_{i}\right)\)…. ( pre-multiply both sides by \(\mathbf{X}\) )
-
\(\frac{1}{N} \mathbf{X} \mathbf{X}^{\mathrm{T}} \mathbf{v}_{i}=\lambda_{i} \mathbf{v}_{i}\) …. ( define \(\mathbf{v}_{i}=\mathbf{X} \mathbf{u}_{i}\) )
\(\rightarrow\) eigenvector equation for \(N \times N\) matrix \(N^{-1} XX^T\)
-
\(\left(\frac{1}{N} \mathbf{X}^{\mathrm{T}} \mathbf{X}\right)\left(\mathbf{X}^{\mathrm{T}} \mathbf{v}_{i}\right)=\lambda_{i}\left(\mathbf{X}^{\mathrm{T}} \mathbf{v}_{i}\right)\).
- \(\left(\mathbf{X}^{\mathrm{T}} \mathbf{v}_{i}\right)\) is an eigenvector of \(\mathbf{S}\) with eigenvalue \(\lambda_{i}\)
-
\(\mathbf{u}_{i}=\frac{1}{\left(N \lambda_{i}\right)^{1 / 2}} \mathbf{X}^{\mathrm{T}} \mathbf{v}_{i}\\\) (after normalization)
(4) Summary
-
from \(O(D^3)\) to \(O(N^3)\)
-
first, evaluate \(XX^T\)
then, find its eigenvectors & eigenvalues
then, compute eigenvectors in the original data space, using \(\mathbf{u}_{i}=\frac{1}{\left(N \lambda_{i}\right)^{1 / 2}} \mathbf{X}^{\mathrm{T}} \mathbf{v}_{i}\\\)
12-2. Probabilsitic PCA (PPCA)
advantages compared with conventional PCA
-
constrained form of the Gaussian distribution
-
derive EM for PCA \(\rightarrow\) computationally efficient
-
deal with missing values
-
mixtures of PPCA can be formulated
-
PPCA form the basis for Bayesian treatment of PCA
( = dimensionality of the principal subspace can be found automatically from the data )
-
can be run generatively
( = sampling from the distribution )
Introduction
- Closely related to “factor analysis”
- example of linear-Gaussian framework
- formulation
- explicit latent variable \(z\) ( = principal component )
- define Gaussian prior over \(z\) ( = \(p(\mathbf{z})=\mathcal{N}(\mathbf{z} \mid \mathbf{0}, \mathbf{I})\) )
- then, \(p(\mathbf{x} \mid \mathbf{z})=\mathcal{N}\left(\mathbf{x} \mid \mathbf{W} \mathbf{z}+\boldsymbol{\mu}, \sigma^{2} \mathbf{I}\right)\)
\(p(\mathbf{x} \mid \mathbf{z})=\mathcal{N}\left(\mathbf{x} \mid \mathbf{W} \mathbf{z}+\boldsymbol{\mu}, \sigma^{2} \mathbf{I}\right)\).
- mean : linear function of \(\mathbf{z}\)
- ( \(W\) : \(D \times M\) matrix )
- ( \(\mu\) : \(D\) dimensional vector )
Generative viewpoint
\(\mathbf{x}=\mathbf{W}\mathbf{z}+\boldsymbol{\mu}+\boldsymbol{\epsilon}\).
- \(z\) : \(M\) dim Gaussian latent variable
- \(\epsilon\) : \(D\) -dim noise variable, with covariance \(\sigma^2 I\)
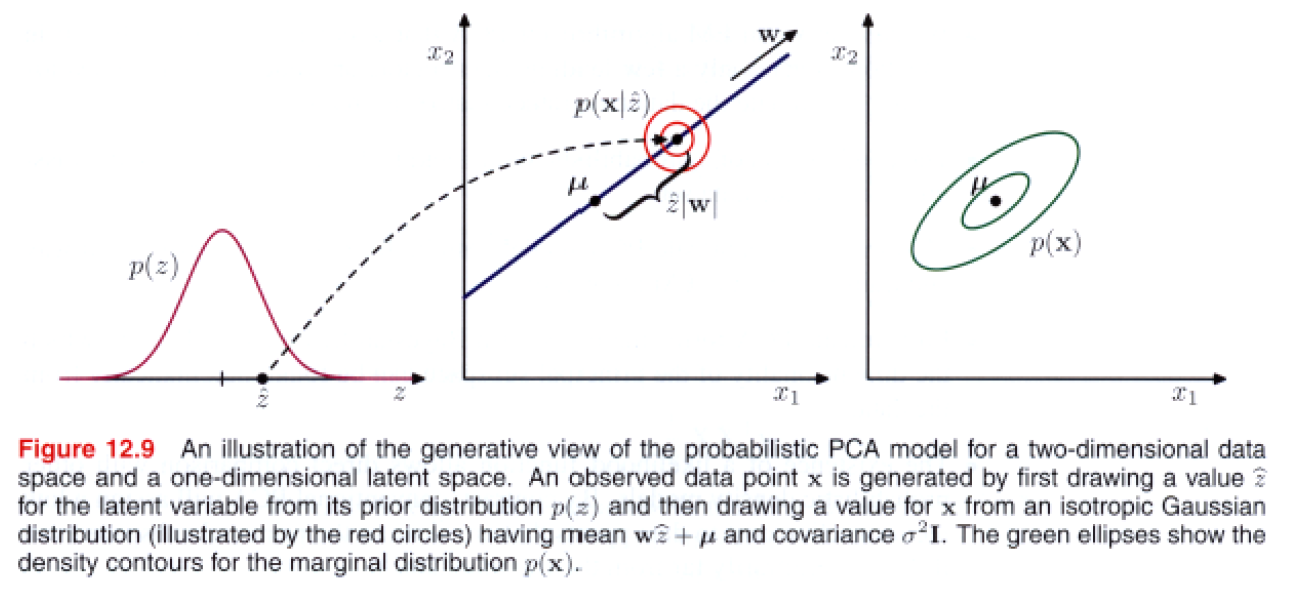
Marginal distribution ( predictive distribution ) \(p(x)\)
How to find parameters \(\mathbf{W}, \boldsymbol{\mu}\) and \(\sigma^{2}\) ?
\(\rightarrow\) likelihood function! need marginal distribution \(p(x)\)
\(p(\mathbf{x})=\int p(\mathbf{x} \mid \mathbf{z}) p(\mathbf{z}) \mathrm{d} \mathbf{z}\).
\(p(\mathbf{x})=\mathcal{N}(\mathbf{x} \mid \boldsymbol{\mu}, \mathbf{C})\).
- where \(\mathbf{C}=\mathbf{W} \mathbf{W}^{\mathrm{T}}+\sigma^{2} \mathbf{I}\)
proof )
\(\begin{aligned} \mathbb{E}[\mathbf{x}] &=\mathbb{E}[\mathbf{W} \mathbf{z}+\boldsymbol{\mu}+\boldsymbol{\epsilon}]=\boldsymbol{\mu} \\ \operatorname{cov}[\mathbf{x}] &=\mathbb{E}\left[(\mathbf{W} \mathbf{z}+\boldsymbol{\epsilon})(\mathbf{W} \mathbf{z}+\boldsymbol{\epsilon})^{\mathrm{T}}\right] \\ &=\mathbb{E}\left[\mathbf{W} \mathbf{z} \mathbf{z}^{\mathrm{T}} \mathbf{W}^{\mathrm{T}}\right]+\mathbb{E}\left[\boldsymbol{\epsilon} \boldsymbol{\epsilon}^{\mathrm{T}}\right]=\mathbf{W} \mathbf{W}^{\mathrm{T}}+\sigma^{2} \mathbf{I} \end{aligned}\).
To evaluate \(p(\mathbf{x})\), we need to evaluate \(C^{-1}\)
- where \(\mathbf{C}=\mathbf{W} \mathbf{W}^{\mathrm{T}}+\sigma^{2} \mathbf{I}\)
-
thus, need inversion of \(D \times D\) matrix
-
can be reduced, using \(\left(\mathbf{A}+\mathbf{B} \mathbf{D}^{-1} \mathbf{C}\right)^{-1}=\mathbf{A}^{-1}-\mathbf{A}^{-1} \mathbf{B}\left(\mathbf{D}+\mathbf{C A}^{-1} \mathbf{B}\right)^{-1} \mathbf{C A}^{-1}\)
\(\rightarrow\) \(\mathrm{C}^{-1}=\sigma^{-1} \mathrm{I}-\sigma^{-2} \mathrm{WM}^{-1} \mathrm{~W}^{\mathrm{T}}\).
where \(\mathbf{M}=\mathbf{W}^{T} \mathbf{W}+\sigma^{2} \mathbf{I}\)
- Thus, \(O(D^3)\) \(\rightarrow\) \(O(M^3)\)
Posterior distribution \(p(z\mid x)\)
\(p(\mathbf{z} \mid \mathbf{x})=\mathcal{N}\left(\mathbf{z} \mid \mathbf{M}^{-1} \mathbf{W}^{\mathrm{T}}(\mathbf{x}-\boldsymbol{\mu}), \sigma^{-2} \mathbf{M}\right)\).
- mean : depends on \(x\)
- cov : independent of \(x\)
12-2-1. Maximum Likelihood PCA
expressed as a directed graph!
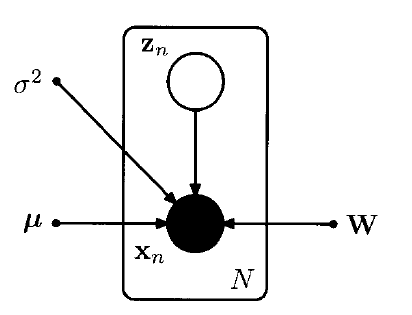
\(\begin{array}{l} \ln p\left(\mathbf{X} \mid \boldsymbol{\mu}, \mathbf{W}, \sigma^{2}\right)&=\sum_{n=1}^{N} \ln p\left(\mathbf{x}_{n} \mid \mathbf{W}, \boldsymbol{\mu}, \sigma^{2}\right) \\ &= -\frac{N D}{2} \ln (2 \pi)-\frac{N}{2} \ln |\mathbf{C}|-\frac{1}{2} \sum_{n=1}^{N}\left(\mathbf{x}_{n}-\boldsymbol{\mu}\right)^{\mathrm{T}} \mathbf{C}^{-1}\left(\mathbf{x}_{n}-\boldsymbol{\mu}\right) \end{array}\).
-
if we solve MLE : \(\mu=\overline{\mathrm{x}}\).
\(\ln p\left(\mathbf{X} \mid \mathbf{W}, \boldsymbol{\mu}, \sigma^{2}\right)=-\frac{N}{2}\left\{D \ln (2 \pi)+\ln \mid \mathbf{C}\mid+\operatorname{Tr}\left(\mathbf{C}^{-1} \mathbf{S}\right)\right\}\).
- \(\mathbf{S}=\frac{1}{N} \sum_{n=1}^{N}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)^{\mathrm{T}}\).
Maximization of \(\ln p\left(\mathbf{X} \mid \boldsymbol{\mu}, \mathbf{W}, \sigma^{2}\right)\)
- \(\mathbf{W}_{\mathrm{ML}}=\mathbf{U}_{M}\left(\mathbf{L}_{M}-\sigma^{2} \mathbf{I}\right)^{1 / 2} \mathbf{R}\).
- \(\mathbf{U}_{M}\) : \(D \times M\) matrix, of subset of the eigenvectors of the cov matrix \(S\)
- \(\mathbf{L}_m\) : \(M \times M\) diagonal matrix ( with eigenvalues \(\lambda_i\) )
- \(\mathbf{R}\) : arbitrary \(M \times M\) orthogonal matrix
- \(\sigma_{\mathrm{ML}}^{2}=\frac{1}{D-M} \sum_{i=M+1}^{D} \lambda_{i}\).
12-2-2. EM algorithm for PCA
PPCA : marginalization over a continuous latent variable \(z\)
( each data point \(x_n\) has its corresponding \(z_n\))
use EM algorithm to find MLE of parameters!
- adv 1) seems awkward.. but in HIGH DIMENSIONAL CASE : computational advantages!
- adv 2) can be extended to factor analysis
- adv 3) allows missing data to be handled
Complete-data log likelihood
\(\ln p\left(\mathbf{X}, \mathbf{Z} \mid \boldsymbol{\mu}, \mathbf{W}, \sigma^{2}\right)=\sum_{n=1}^{N}\left\{\ln p\left(\mathbf{x}_{n} \mid \mathbf{z}_{n}\right)+\ln p\left(\mathbf{z}_{n}\right)\right\}\).
\(\begin{aligned} \mathbb{E}[\ln p&\left.\left(\mathbf{X}, \mathbf{Z} \mid \boldsymbol{\mu}, \mathbf{W}, \sigma^{2}\right)\right]=-\sum_{n=1}^{N}\left\{\frac{D}{2} \ln \left(2 \pi \sigma^{2}\right)+\frac{1}{2} \operatorname{Tr}\left(\mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right]\right)\right.\\ &+\frac{1}{2 \sigma^{2}}\left\|\mathbf{x}_{n}-\boldsymbol{\mu}\right\|^{2}-\frac{1}{\sigma^{2}} \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}} \mathbf{W}^{\mathrm{T}}\left(\mathbf{x}_{n}-\boldsymbol{\mu}\right) \\ &\left.+\frac{1}{2 \sigma^{2}} \operatorname{Tr}\left(\mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right] \mathbf{W}^{\mathrm{T}} \mathbf{W}\right)\right\} \end{aligned}\).
E-step
use old parameter values to evaluate
- \(\mathbb{E}\left[\mathbf{z}_{n}\right] =\mathbf{M}^{-1} \mathbf{W}^{\mathrm{T}}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\).
- \(\mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right] =\sigma^{2} \mathbf{M}^{-1}+\mathbb{E}\left[\mathbf{z}_{n}\right] \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}}\).
M-step
maximize w.r.t \(\mathbf{W}\) and \(\sigma^2\)
\[\begin{aligned} \mathbf{W}_{\text {new }}=&\left[\sum_{n=1}^{N}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right) \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}}\right]\left[\sum_{n=1}^{N} \mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right]\right]^{-1} \\ \sigma_{\text {new }}^{2}=& \frac{1}{N D} \sum_{n=1}^{N}\left\{\left\|\mathbf{x}_{n}-\overline{\mathbf{x}}\right\|^{2}-2 \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}} \mathbf{W}_{\text {new }}^{\mathrm{T}}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\right.\\ &\left.+\operatorname{Tr}\left(\mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right] \mathbf{W}_{\text {new }}^{\mathrm{T}} \mathbf{W}_{\text {new }}\right)\right\} \end{aligned}\]Computational Efficiency
- eigen decomposition of
- covariance matrix : \(O(D^3)\)
- interested in \(M\) ev : \(O(MD^2)\)
-
evaluation of covariance matrix : \(O(ND^2)\)
( \(N\) : number of data points )
-
use “snapshot method”
- eigenvectors are linear combinations of data vectors
- avoid evaluation of covariance matrix
- but \(O(N^3)\) ( not appropriate for large datasets )
-
EM algorithm
-
do not construct covariance matrix explicitly
-
\(O(NDM)\).
( can be appropriate for large \(D\) and \(M<<D\) )
-
Others
-
Deal with missing data
( by marginalizing over the distribution of unobserved)
-
take limit \(\sigma^2 \rightarrow 0\), corresponding to standard PCA
12-2-3. Bayesian PCA
Seek Bayesian approach to “model selection”
- Can be compared to different \(M\)
\(\rightarrow\) need to marginalize out \(\mu, \mathrm{W}, \text { and } \sigma^{2}\)
Evidence Approximation
-
choice of prior over \(\mathbf{W}\)
( allows surplus dimensions in the principal subspace to be pruned out)
\(\rightarrow\) ARD (Automatic Relevance Determination)
ARD (Automatic Relevance Determination)
- each Gaussian has independent variance, governed by hyperparameter \(\alpha_i\)
- \(p(\mathbf{W} \mid \boldsymbol{\alpha})=\prod_{i=1}^{M}\left(\frac{\alpha_{i}}{2 \pi}\right)^{D / 2} \exp \left\{-\frac{1}{2} \alpha_{i} \mathbf{w}_{i}^{\mathrm{T}} \mathbf{w}_{i}\right\}\).
-
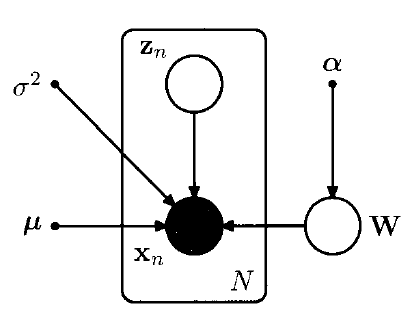
-
\(\alpha_i \rightarrow \infty\) = \(w_i \rightarrow 0\)
-
“effective dimensionality of the principal subspace is then determined by the number of finite \(\alpha_i\) values,
and the corresponding vectors \(w_i\) can be thought of as ‘relevant’ for modeling the data distribution
Estimation of \(\alpha\)
-
\(\alpha_i\) are re-estimated during training
-
by maximizing the log marginal likelihood :
\(p\left(\mathbf{X} \mid \boldsymbol{\alpha}, \boldsymbol{\mu}, \sigma^{2}\right)=\int p\left(\mathbf{X} \mid \mathbf{W}, \boldsymbol{\mu}, \sigma^{2}\right) p(\mathbf{W} \mid \boldsymbol{\alpha}) \mathrm{d} \mathbf{W}\).
-
intractable! use Laplace approximation
-
Result : \(\alpha_{i}^{\text {new }}=\frac{D}{\mathbf{w}_{i}^{\mathrm{T}} \mathbf{w}_{i}}\)
E-step
use old parameter values to evaluate
- \(\mathbb{E}\left[\mathbf{z}_{n}\right] =\mathbf{M}^{-1} \mathbf{W}^{\mathrm{T}}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\).
- \(\mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right] =\sigma^{2} \mathbf{M}^{-1}+\mathbb{E}\left[\mathbf{z}_{n}\right] \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}}\).
M-step
maximize w.r.t \(\mathbf{W}\) and \(\sigma^2\)
\(\begin{aligned} \mathbf{W}_{\text {new }}&=\left[\sum_{n=1}^{N}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right) \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}}\right]\left[\sum_{n=1}^{N} \mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right]+\sigma^{2} \mathbf{A}\right]^{-1} \\ \sigma_{\text {new }}^{2}=& \frac{1}{N D} \sum_{n=1}^{N}\left\{\left\|\mathbf{x}_{n}-\overline{\mathbf{x}}\right\|^{2}-2 \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}} \mathbf{W}_{\text {new }}^{\mathrm{T}}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)\right.\\ &\left.+\operatorname{Tr}\left(\mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right] \mathbf{W}_{\text {new }}^{\mathrm{T}} \mathbf{W}_{\text {new }}\right)\right\} \end{aligned}\).
where \(\mathbf{A}=\operatorname{diag}\left(\alpha_{i}\right) .\)
12-2-4. Factor Analysis
-
linear-Gaussian latent variable model
-
closely related to PPCA
( difference ) conditional distribution : diagonal covariance
\(p(\mathbf{x} \mid \mathbf{z})=\mathcal{N}(\mathbf{x} \mid \mathbf{W} \mathbf{z}+\boldsymbol{\mu}, \boldsymbol{\Psi})\).
- where \(\Psi\) is a \(D \times D\) diagonal matrix
“factor loadings”
-
Columns of \(\mathbf{W}\)
-
capture the correlation between observation variables
“uniqueness”
- diagonal elements of \(\mathbf{\Psi}\)
- represent the independent noise variance
Marginal distribution for the observed variable
\(p(\mathbf{x})=\mathcal{N}(\mathbf{x} \mid \boldsymbol{\mu}, \mathbf{C})\).
-
where \(\mathbf{C}=\mathbf{W} \mathbf{W}^{\mathrm{T}}+\boldsymbol{\Psi}\)
( PPCA : \(\mathbf{C}=\mathbf{W} \mathbf{W}^{\mathrm{T}}+\sigma^{2} \mathbf{I}\) )
-
like PPCA, model is invariant to the rotations in the latent space
Unlike PPCA, there is no longer closed form!
\(\rightarrow\) use EM algorithm
E-step
\(\begin{aligned} \mathbb{E}\left[\mathbf{z}_{n}\right] &=\mathbf{G} \mathbf{W}^{\mathrm{T}} \boldsymbol{\Psi}^{-1}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right) \\ \mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right] &=\mathbf{G}+\mathbb{E}\left[\mathbf{z}_{n}\right] \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}} \end{aligned}\).
where \(\mathbf{G}=\left(\mathbf{I}+\mathbf{W}^{\mathrm{T}} \boldsymbol{\Psi}^{-1} \mathbf{W}\right)^{-1}\).
- need inversion of \(M \times M\) matrices ( not \(D \times D\))
M-step
\(\begin{aligned} \mathbf{W}^{\text {new }} &=\left[\sum_{n=1}^{N}\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right) \mathbb{E}\left[\mathbf{z}_{n}\right]^{\mathrm{T}}\right]\left[\sum_{n=1}^{N} \mathbb{E}\left[\mathbf{z}_{n} \mathbf{z}_{n}^{\mathrm{T}}\right]\right]^{-1} \\ \boldsymbol{\Psi}^{\text {new }} &=\operatorname{diag}\left\{\mathbf{S}-\mathbf{W}_{\text {new }} \frac{1}{N} \sum_{n=1}^{N} \mathbb{E}\left[\mathbf{z}_{n}\right]\left(\mathbf{x}_{n}-\overline{\mathbf{x}}\right)^{\mathrm{T}}\right\} \end{aligned}\)/
12-3. Kernel PCA
replacing the scalar products with NON-linear kernel
Review
- PCs are defined by eigenvectors \(u_i\) of covariance matrix \(S\)
- \(\mathbf{S u}_{i}=\lambda_{i} \mathbf{u}_{i}\).
- \(\mathbf{S}=\frac{1}{N} \sum_{n=1}^{N} \mathbf{x}_{n} \mathbf{x}_{n}^{\mathrm{T}}\) : \(D \times D\) sample covariance matrix
- \(\mathbf{u}_{i}^{\mathrm{T}} \mathbf{u}_{i}=1\) : normalized eigenvector
Nonlinear transformation \(\phi(x)\)
-
perform standard PCA in the “feature space”
( = “nonlinear” PC model in the “original model”)
-
\(\mathrm{Cv}_{i}=\lambda_{i} \mathbf{v}_{i}\).
- \(\mathbf{C}=\frac{1}{N} \sum_{n=1}^{N} \phi\left(\mathbf{x}_{n}\right) \phi\left(\mathbf{x}_{n}\right)^{\mathrm{T}}\) : \(M \times M\) sample covariance matrix
- \(\lambda_{i} \mathbf{v}_{i}= \frac{1}{N} \sum_{n=1}^{N} \phi\left(\mathbf{x}_{n}\right)\left\{\phi\left(\mathbf{x}_{n}\right)^{\mathrm{T}} \mathbf{v}_{i}\right\}\).
- \(\mathbf{v}_{i}=\sum_{n=1}^{N} a_{i n} \phi\left(\mathbf{x}_{n}\right)\).
[ Eigenvector equation ]
\(\mathrm{Cv}_{i}=\lambda_{i} \mathbf{v}_{i}\).
\(\frac{1}{N} \sum_{n=1}^{N} \phi\left(\mathbf{x}_{n}\right) \phi\left(\mathbf{x}_{n}\right)^{\mathrm{T}} \sum_{m=1}^{N} a_{i m} \phi\left(\mathbf{x}_{m}\right)=\lambda_{i} \sum_{n=1}^{N} a_{i n} \phi\left(\mathbf{x}_{n}\right)\).
( since \(k\left(\mathbf{x}_{n}, \mathbf{x}_{m}\right)=\phi\left(\mathbf{x}_{n}\right)^{\mathrm{T}} \phi\left(\mathrm{x}_{m}\right)\) )
\(\frac{1}{N} \sum_{n=1}^{N} k\left(\mathbf{x}_{l}, \mathbf{x}_{n}\right) \sum_{m=1}^{m} a_{i m} k\left(\mathbf{x}_{n}, \mathbf{x}_{m}\right)=\lambda_{i} \sum_{n=1}^{N} a_{i n} k\left(\mathbf{x}_{l}, \mathbf{x}_{n}\right)\).
( as matrix notation )
\(\mathbf{K}^{2} \mathbf{a}_{i}=\lambda_{i} N \mathbf{K} \mathbf{a}_{i}\).
\(\mathbf{K} \mathbf{a}_{i}=\lambda_{i} N \mathbf{a}_{i}\).
Normalization condition is obtained!
-
proof )
\(1=\mathbf{v}_{i}^{\mathrm{T}} \mathbf{v}_{i}=\sum_{n=1}^{N} \sum_{m=1}^{N} a_{i n} a_{i m} \phi\left(\mathbf{x}_{n}\right)^{\mathrm{T}} \boldsymbol{\phi}\left(\mathbf{x}_{m}\right)=\mathbf{a}_{i}^{\mathrm{T}} \mathbf{K} \mathbf{a}_{i}=\lambda_{i} N \mathbf{a}_{i}^{\mathrm{T}} \mathbf{a}_{i}\).
Projection of point \(\mathbf{x}\) onto eigenvector \(i\):
- \(y_{i}(\mathbf{x})=\boldsymbol{\phi}(\mathbf{x})^{\mathrm{T}} \mathbf{v}_{i}=\sum_{n=1}^{N} a_{i n} \boldsymbol{\phi}(\mathbf{x})^{\mathrm{T}} \boldsymbol{\phi}\left(\mathbf{x}_{n}\right)=\sum_{n=1}^{N} a_{i n} k\left(\mathbf{x}, \mathbf{x}_{n}\right)\).
Original PCA vs Kernel PCA
-
original) can have at most \(D\) PCs
-
kernel) can exceed \(D\) PCs
( but cannot exceed \(N\), which is the number of data points, since covariance matrix in feature space has rank at most equal to \(N\) )
Centralize
-
cannot simply compute and then subtract off the mean
-
should be done “in terms of the kernel function”
\(\widetilde{\phi}\left(\mathbf{x}_{n}\right)=\phi\left(\mathbf{x}_{n}\right)-\frac{1}{N} \sum_{l=1}^{N} \phi\left(\mathbf{x}_{l}\right)\).
Gram matrix : \(\widetilde{\mathbf{K}}=\mathbf{K}-\mathbf{1}_{N} \mathbf{K}-\mathbf{K} \mathbf{1}_{N}+\mathbf{1}_{N} \mathbf{K} \mathbf{1}_{N}\)
Example )
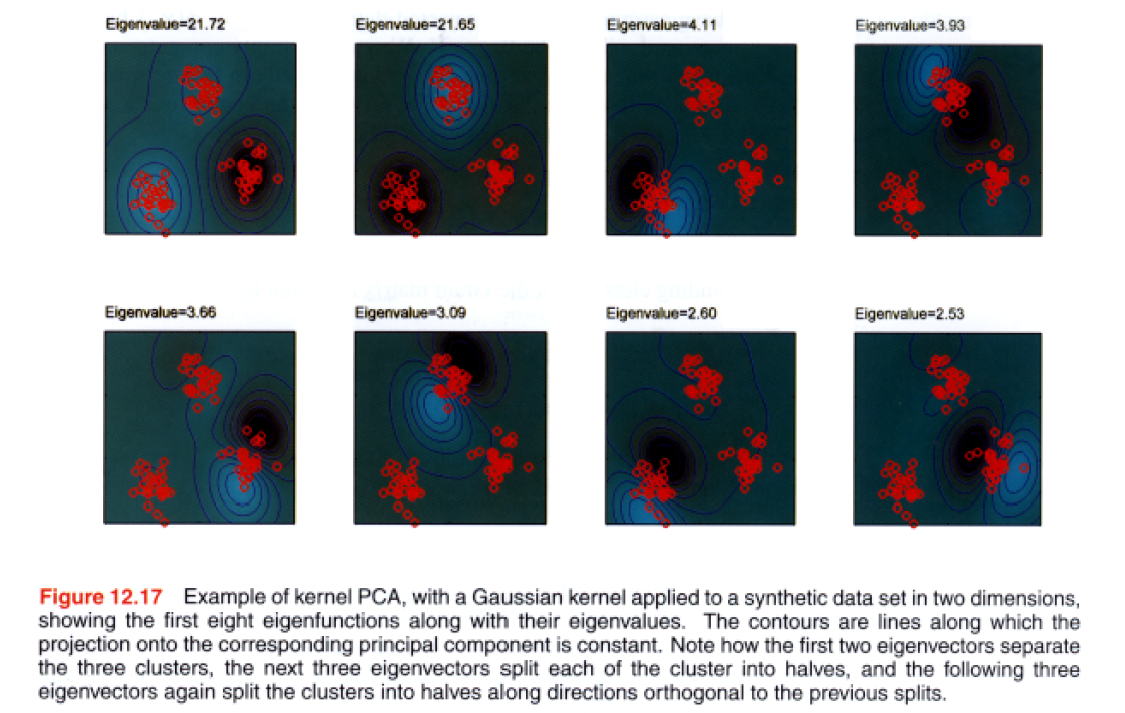
-
Gaussian’ kernel : \(k\left(\mathbf{x}, \mathbf{x}^{\prime}\right)=\exp \left(-\left\|\mathbf{x}-\mathbf{x}^{\prime}\right\|^{2} / 0.1\right)\)
-
lines correspond to contours,
\(\phi(\mathbf{x})^{\mathrm{T}} \mathbf{v}_{i}=\sum^{N} a_{i n} k\left(\mathbf{x}, \mathbf{x}_{n}\right)\).
Disadvantage of Kernel PCA
-
involves finding eigenvector of \(N \times N\) matrix \(\tilde{\mathbf{K}}\) ( not \(D \times D\) matrix \(S\) )
\(\rightarrow\) for large datasets, approximation is needed
-
standard PCA ) we can approximate a data vector \(x_n\) by \(\hat{x_n}\)
like \(\widehat{\mathbf{x}}_{n}=\sum_{i=1}^{L}\left(\mathbf{x}_{n}^{\mathrm{T}} \mathbf{u}_{i}\right) \mathbf{u}_{i}\).
kernel PCA) not possible
12-4. Nonlinear Latent Variable Models
generalization of this framework to “nonlinear” or “non-Gaussian”
12-4-1. ICA (Independent Component Analysis)
Framework
-
observed variables are related linearly to the latent variables
-
latent distribution is “NON-GAUSSIAN”
\(p(\mathbf{z})=\prod_{j=1}^{M} p\left(z_{j}\right)\).
12-4-2. Autoassociative Neural Networks
NN has been used for dimensionality reduction
minimize some measure of the reconstruction error
- \(E(\mathbf{w})=\frac{1}{2} \sum_{n=1}^{N}\left\|\mathbf{y}\left(\mathbf{x}_{n}, \mathbf{w}\right)-\mathbf{x}_{n}\right\|^{2}\).
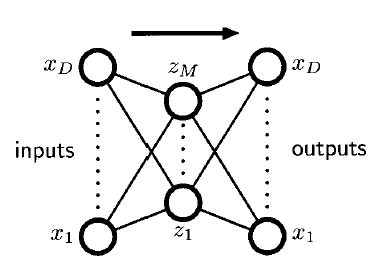
- two layers of weights
- equivalent to linear PCA
Limitations of a linear dimensionality could be overcome by NON-LINEAR activation functions
( additional hidden layers are permitted ! )
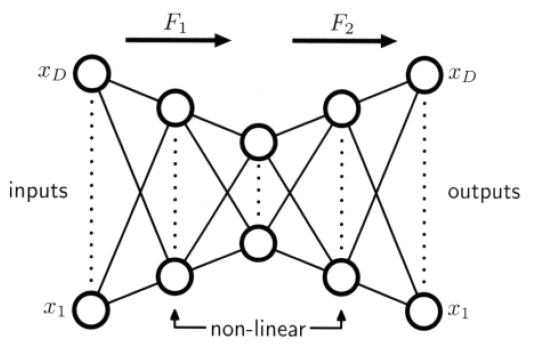
- four layer
- sigmoidal nonlinear activation
- not being limited to linear transformations!
- nonlinear optimization problem
