[ 15. Deep Generative Models for Graphs ]
( 참고 : CS224W: Machine Learning with Graphs )
Contents
- ML for Graph Generation
- GraphRNN : Generating Realistic Graphs
- BFS (Breadth-First Search node ordering)
- Evaluating Generated Graphs
- Application of Deep Graph Generative Models
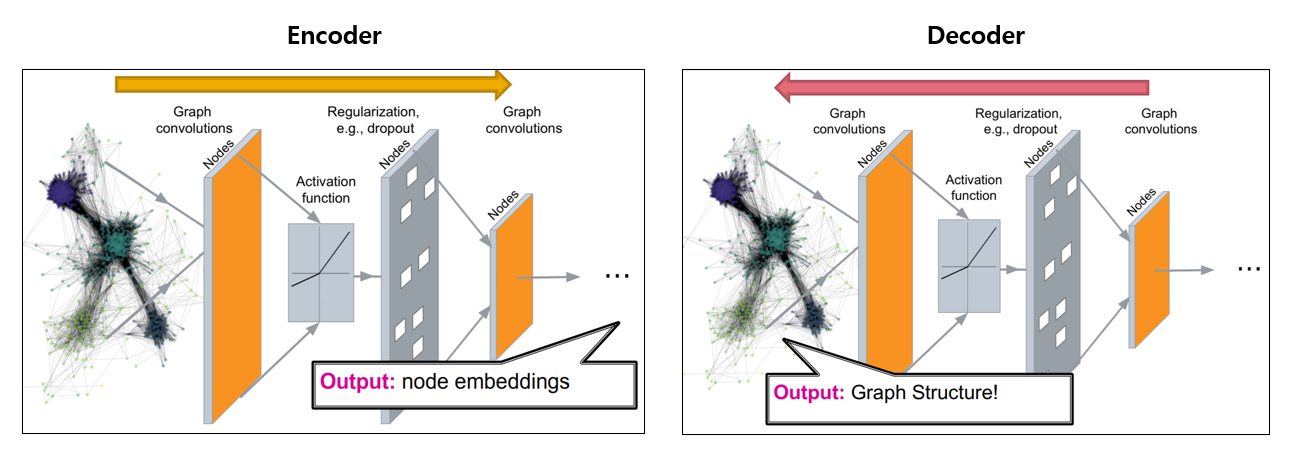
Deep graph encoders & DECODERS
1. ML for Graph Generation
(1) Tasks
[ Task 1 ] Realistic graph generation
- generate graph, that are “similar to a given set of graphs”
[ Task 2 ] Goal-directed graph generation
- for specific objective! ( with constraints )
(2) Graph Generative Models
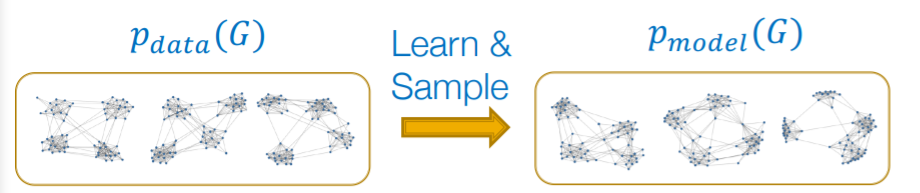
Given : graphs \(\{x_i\}\) ( sampled from \(p_{data}(G)\) )
- we do not have true distribution, but only some samples of it
Goal
- goal 1) learn \(p_{model}(G)\)
- goal 2) sample from \(p_{model}(G)\)
(3) Basics for generative models
Notation
- \(p_{data}(x)\) : data distribution
- unknown, but have some samples of it ( \(x_i \sim p_{data}(x)\) )
- \(p_{model}(x ; \theta)\) : model
- use this to approximate \(p_{data}(x)\)
Goal
- Density estimation
- make \(p_{model}(x ; \theta)\) close to \(p_{data}(x)\)
- Sampling
- \[x_i \sim p_{model}(x ; \theta)\]
a) Density estimation
key point : MLE
- \(\boldsymbol{\theta}^{*}=\underset{\boldsymbol{\theta}}{\arg \max } \mathbb{E}_{x \sim p_{\text {data }}} \log p_{\text {model }}(\boldsymbol{x} \mid \boldsymbol{\theta})\).
- finding a model, that is most likely to have generated the observed \(x\)
b) Sampling
goal : sample from a “complex distribution”
most common approach
-
step 1) sample from noise : \(\boldsymbol{z}_{i} \sim N(0,1)\)
-
step 2) transform the noise : \(x_{i}=f\left(z_{i} ; \theta\right)\)
\(\rightarrow\) key point : use \(f(\cdot)\) with DNN
(4) Deep Generative Models
Auto-regressive models
use “chain rule”
-
\(p_{\text {model }}(\boldsymbol{x} ; \theta)=\prod_{t=1}^{n} p_{\text {model }}\left(x_{t} \mid x_{1}, \ldots, x_{t-1} ; \theta\right)\).
-
(for GNN) \(x_t\) : \(t\)-th action
- ex) adding NODE, adding EDGE
2. GraphRNN : Generating Realistic Graphs
(1) Basic Idea
generate graphs, by sequentially adding nodes/edges
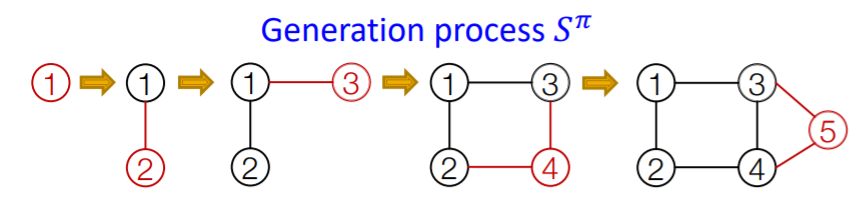
(2) Model graphs as “SEQUENCES”
Notation
- \(G\) : graph
- \(\pi\) : node ordering
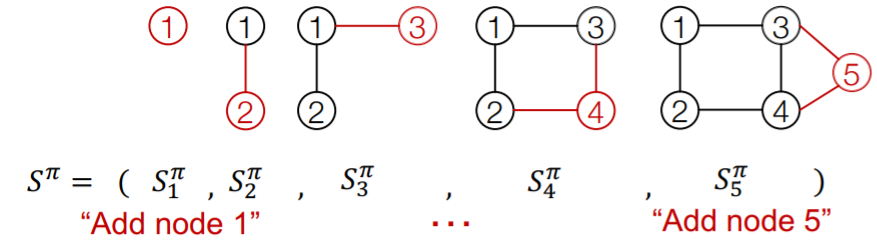
- \(S^{\pi}\) : Sequence
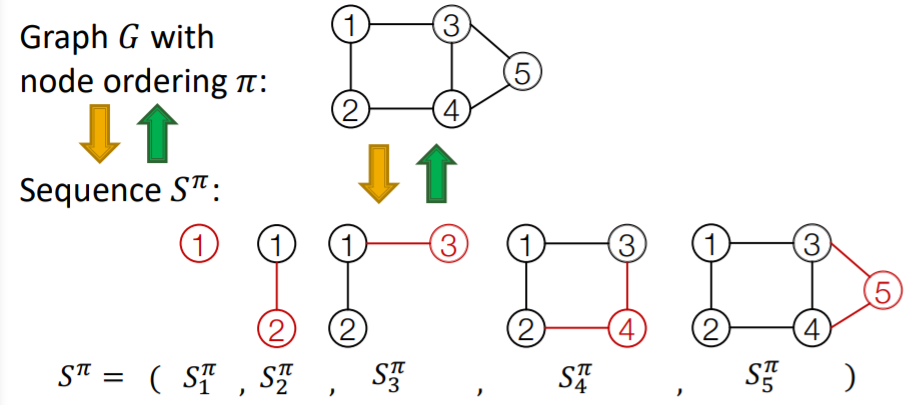
Sequence \(S^{\pi}\) : consists of “2-level”
- 1) node-level : add one node
- 2) edge-level : add edge between existing nodes
each “node-level” step is an “edge-level sequence”
- node-level (a)
- edge-level (a-1)
- edge-level (a-2)
- ..
- node-level (b)
- edge-level (b-1)
- edge-level (b-2)
- ..
Ex) node-level
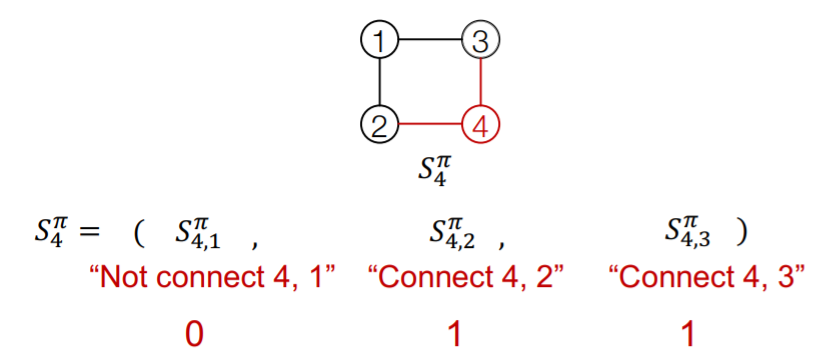
Ex) edge-level
Summary : graph + node ordering = sequence of sequence
- \(\therefore\) graph generation = sequence generation
- ( node ordering : select randomly )
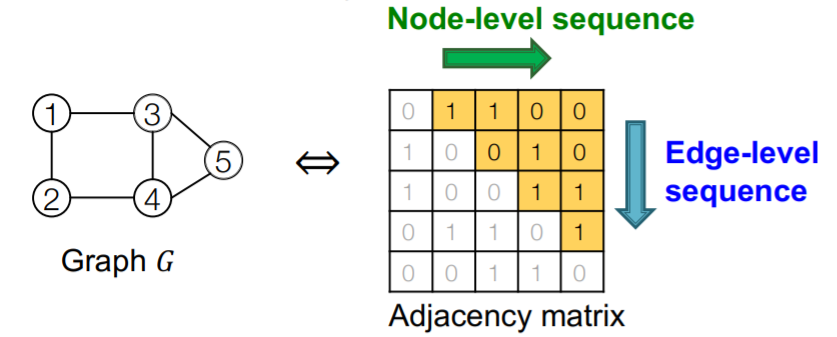
Process : need to model 2 processes
-
1) node-level sequence
- generate a “state for a new node”
-
2) edge-level sequence
- generate edges for “new node”, based on its state
\(\rightarrow\) use RNN for these 2 processes
(3) Graph RNN : 2-level RNN
a) 2-level RNN
- NODE-level RNN ( nRNN )
- EDGE-level RNN ( eRNN )
Relation between 2 RNNs :
- “nRNN” generates the initial state for “eRNN”
- “eRNN” sequentially predicts if the new node will connect to each of the previous node
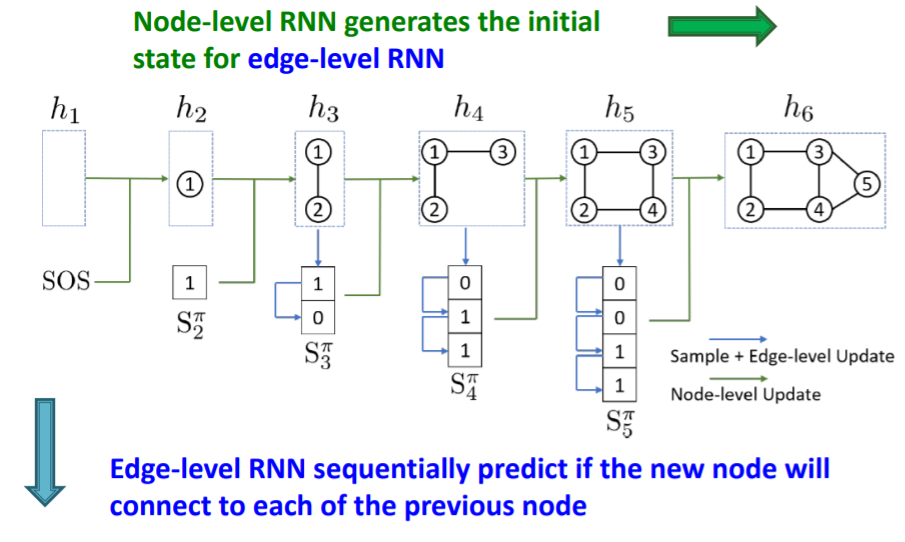
b) key questions
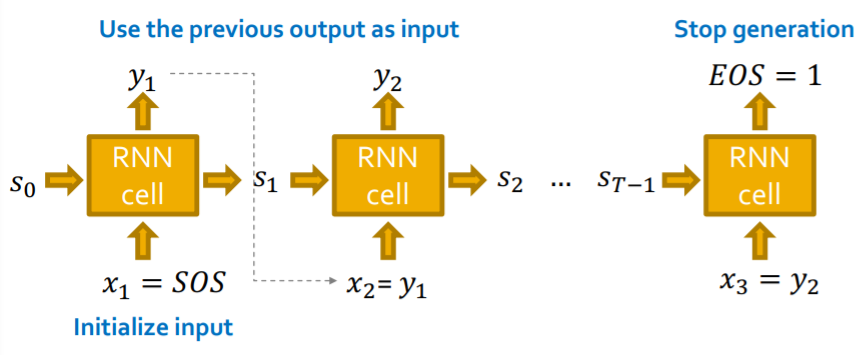
Q1) how does RNN generate sequences?
\(\rightarrow\) let \(x_{t+1} = y_t\) ( input = previous output )
Q2) how to initialize input sequence
\(\rightarrow\) uses SOS token ( zero/one vector )
Q3) when to stop generation?
\(\rightarrow\) use EOS token as extra RNN output
c) deterministic & stochastic
Deterministic output
Stochastic output
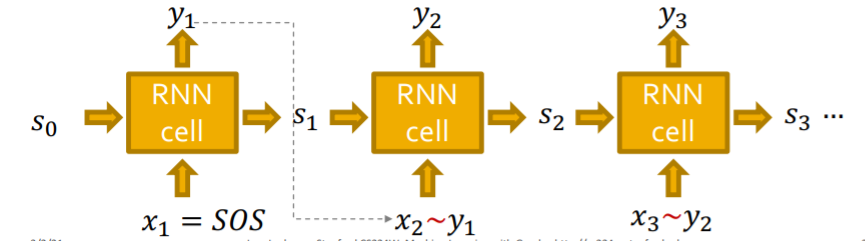
Autoregressive Model ( with RNN )
- \(\prod_{k=1}^{n} p_{\operatorname{model}}\left(x_{t} \mid x_{1}, \ldots, x_{t-1} ; \theta\right)\).
- notation : \(y_{t}=p_{\text {model }}\left(x_{t} \mid x_{1}, \ldots, x_{t-1} ; \theta\right)\)
- sampling : \(y_{t}: x_{t+1} \sim y_{t}\)
- how to make it stochastic?
- output of RNN = “probability of an edge”
- (1) sample from that distribution & (2) feed to next step
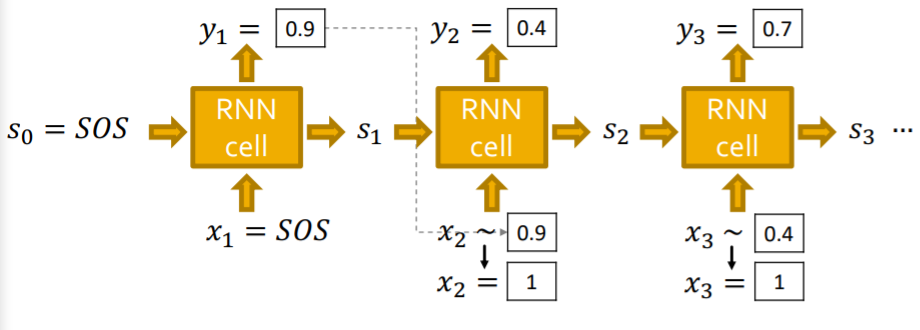
d) RNN at Test Time
Notation
- \(y_t\) : output of RNN at time \(t\)
- follows “Bernoulli distn”
- \(p\) : probability of an edge
- (value=1 of prob \(p\)) & (value=0 of prob \(1-p\))
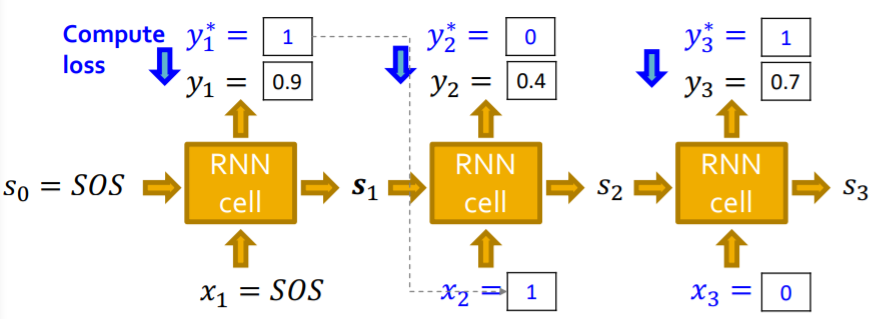
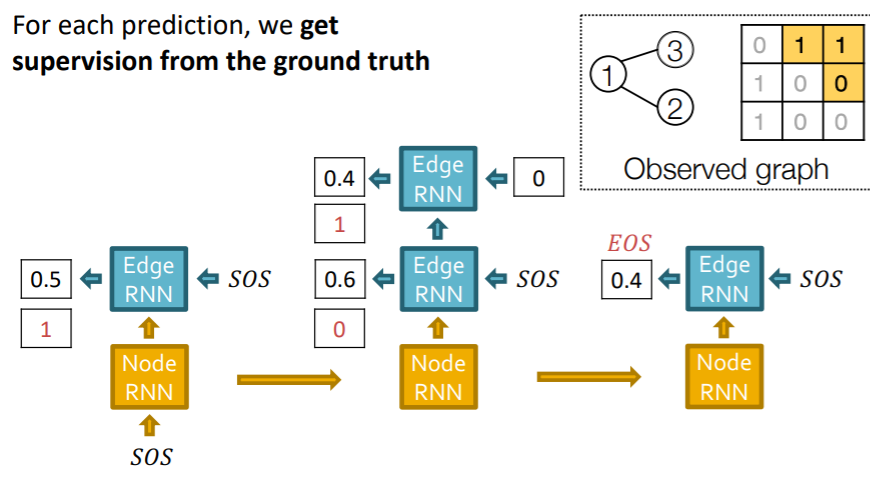
e) RNN at Training Time
observed data (given)
- sequence \(y^{*}\) of edges ( 1 or 0 )
use “Teacher forcing”
- use the “TRUE value”, while training
- loss function : \(L=-\left[y_{1}^{*} \log \left(y_{1}\right)+\left(1-y_{1}^{*}\right) \log \left(1-y_{1}\right)\right]\)
f) Summary
-
step 1) add a new node
-
run Node RNN
( output of Node RNN = initialize Edge RNN )
-
-
step 2) add new edges ( for new node )
-
run Edge RNN
( predict if “new node” connect to “previous node” )
-
-
step 3) add a new node
-
run Node RNN
( use last hidden state of Edge RNN )
-
-
step 4) stop generation
- stop, if Edge RNN outputs EOS at step 1
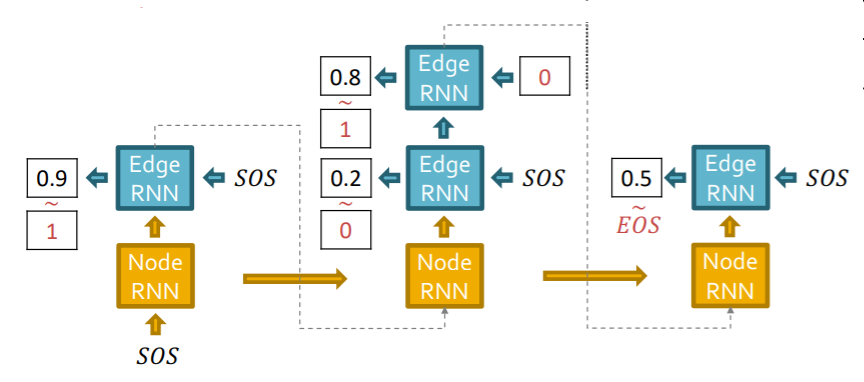
Test time
( just replace “input” at each step, with “GraphRNN’s own predictions” )
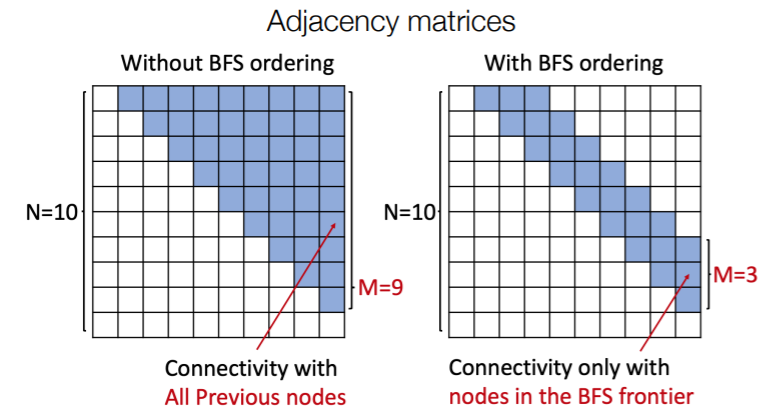
3. BFS (Breadth-First Search node ordering)
How to get tractability?
too many steps are need for edge generation
- problem 1) need “FULL” adjacency matrix
- problem 2) complex “TOO-LONG” edge dependencies
\(\rightarrow\) Solution : BFS (Breadth-First Search node ordering)
BFS (Breadth-First Search node ordering)

if node \(T\) doesn’t connect to node \(K\) ( \(K<T\) ),
then node \(T+\alpha\) doesn’t ~
( neighbors of \(K\) have already been traversed! )
\(\rightarrow\) only need memory of “2 steps” ( not n-1 steps )
With & Without BFS
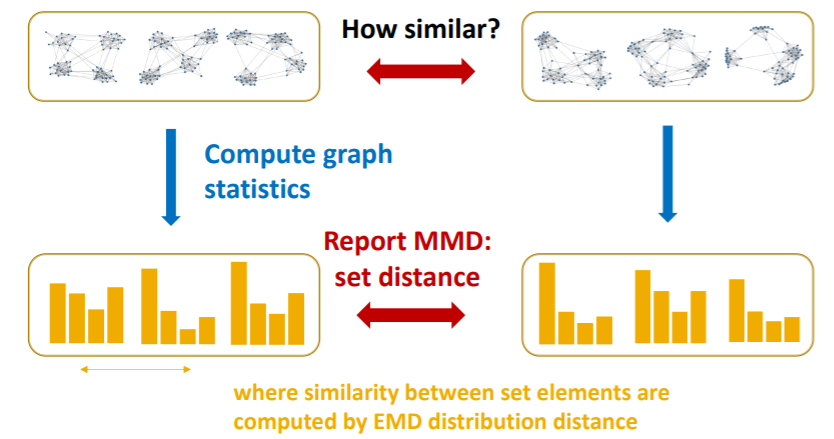
4. Evaluating Generated Graphs
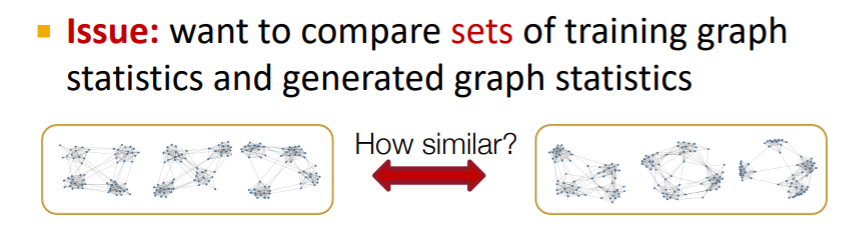
compare 2 sets of graphs
\(\rightarrow\) how much are they similar? need to define “SIMILARITY METRIC”
Solution
- 1) visual similarity
- 2) graph statistics similarity
(1) Visual similarity
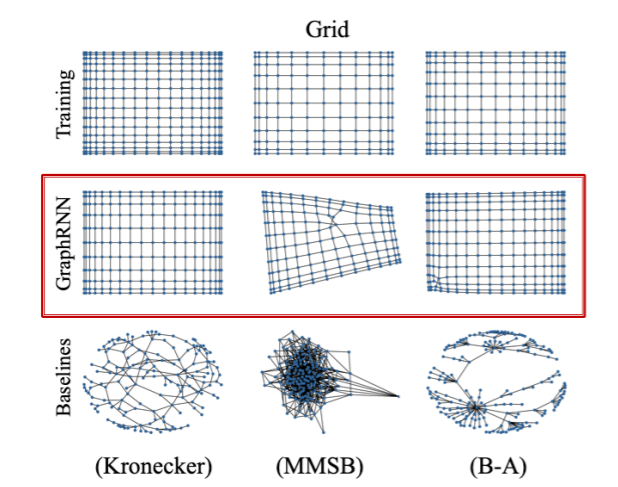
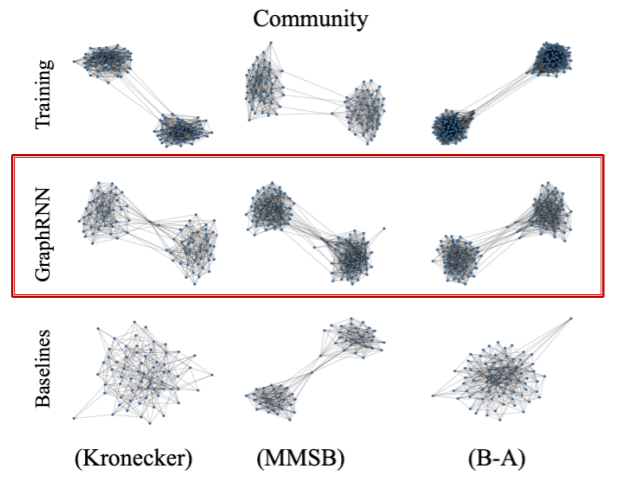
(2) Graph statistics similarity
Typical graph statistics
- 1) degree distribution
- 2) clustering coefficient distribution
- 3) orbit count statistics
\(\rightarrow\) each statistic is “not a scalar”, but “PROBABILITY DISTN”
Solution
- step 1) compare “2 graph statistics”
- by EMD (Earth Mover Distance)
- step 2) compare “sets of graph statistics”
- by MMD (Maximum Mean Discrepancy)
a) EMD
- similarity between “2 distns”
- “minimum effort” that move earth from “one pile to another pile”
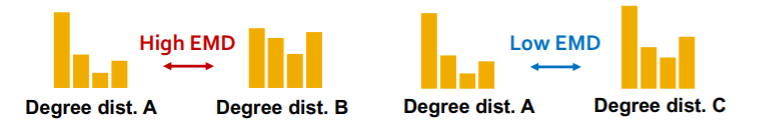
b) MMD
-
similarity between “2 sets”,
based on the similarity between “set elements”

we will compare “2 sets of graph statistics”
( = “2 sets of distributions” )
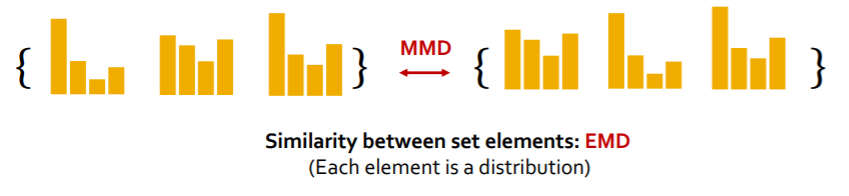
Summary
- step 1) compute “graph statistics” of all graphs
- step 2) calculate “MMD” ( distance between sets )
5. Application of Deep Graph Generative Models
pass
