DPSOM : Deep Probabilistic Clustering with Self-Organizing Maps (2020)
Contents
- Abstract
- Introduction
- Probabilistic clustering with DPSOM
- Background
- PSOM : Probabilistic SOM clustering
- DPSOM : VAE for representation learning
- T-DPSOM : Extension to time series data
0. Abstract
interpretable visualizations from complex data :
- 1) clustering
- 2) representation learning
however, current methods do not combine those well!
present a novel way to ..
- (a) fit SOM with probabilistic cluster assignments ( = PSOM )
- (b) new deep architecture for probabilistic clustering ( = DPSOM )
- (c) extend our architecture for ts clustering ( = T-DPSOM )
1. Introduction
[1] k-means, GMM
- constrained to simple data
- limited in high-dim / complex / real world data
[2] AE, VAE, GAN
- have been used in combination with clustering methods
- compressed latent representation : ease clustering process!
[3] SOM (Self-Organizing Map)
- interpretable representation
- low-dim ( usualy 2d ), discretized representations
- performs poorly on complex high-dim data
- effective for data viz / but few have been combined with DNN
to address issues in [3] SOM,
\(\rightarrow\) propose PSOM / DPSOM / T-DPSOM
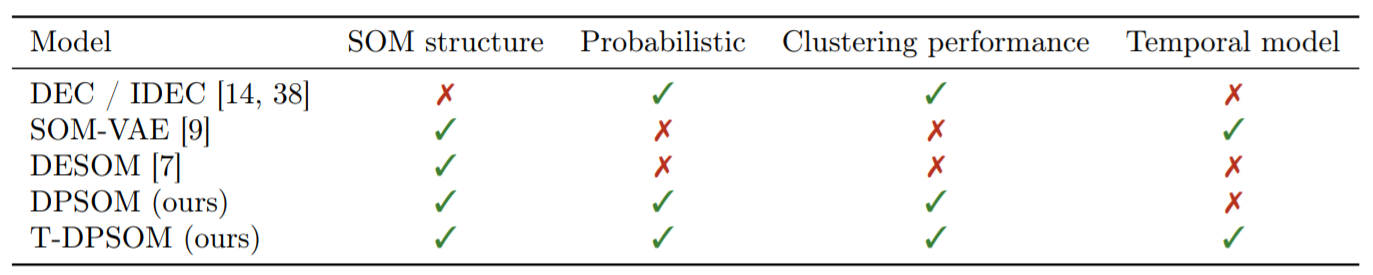
2. Probabilistic clustering with DPSOM
Notation
- data samples \(\left\{x_{i}\right\}_{i=1, \ldots, N}\), where \(x_{i} \in \mathbb{R}^{d}\)
- goal : partition the data into a set of clusters \(\left\{S_{j}\right\}_{j=1, \ldots, K}\)
Proposed architecture
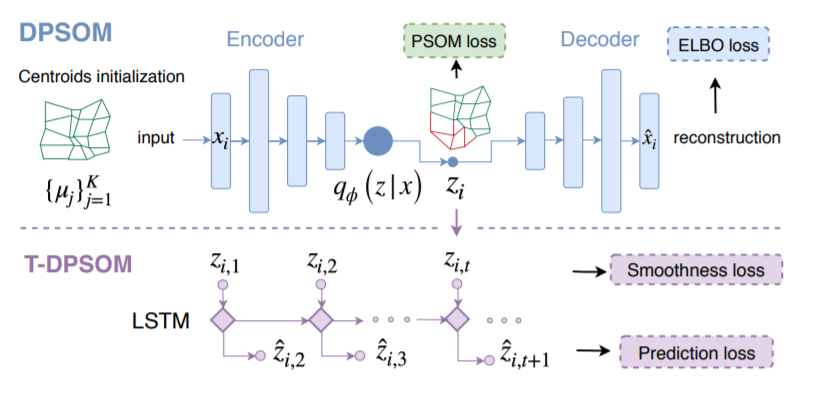
[ DPSOM ]
- use VAE to \(x \rightarrow z\)
- use PSOM to cluster \(z\)
- VAE & PSOM are trained jointly
[ T-DPSOM ]
-
\(z_{i,,t}\) for \(t=1,..,T\) are connected by LSTM
( predict embedding \(z_{t+1}\) )
2-1. Background
SOM is comprised of \(K\) nodes : \(M=\left\{m_{j}\right\}_{j=1}^{K}\)
- node \(m_{j}\) corresponds to a centroid \(\mu_{j}\) in the input space
Algorithm
- randomly selects an input \(x_i\)
- updates both its closest centroid \(\mu_j\) & its neighbors, to move closer to \(x_i\)
CAH ( Clustering Assignment Hardening )
- introduced by DEC model
- perform well in the latent space of AEs
- given an embedding function \(z_i = f(x_i)\), use Student’s t-distn (\(S\)) as a kernel to measure the similarity between \(z_i\) and centroid \(\mu_j\)
- improves cluster purity, by forcing \(S\) to approach a target distn \(T\)
- \(s_{i j}=\frac{\left(1+ \mid \mid z_{i}-\mu_{j} \mid \mid ^{2} / \alpha\right)^{-\frac{\alpha+1}{2}}}{\sum_{j^{\prime}}\left(1+ \mid \mid z_{i}-\mu_{j^{\prime}} \mid \mid ^{2} / \alpha\right)^{-\frac{\alpha+1}{2}}}\).
- \(t_{i j}=\frac{s_{i j}^{\kappa} / \sum_{i^{\prime}} s_{i^{\prime} j}}{\sum_{j^{\prime}} s_{i j^{\prime}}^{\kappa} / \sum_{i^{\prime}} s_{i^{\prime} j^{\prime}}}\).
- clustering loss :
- \(\mathcal{L}_{\mathrm{CAH}}=K L(T \mid \mid S)=\sum_{i=1}^{N} \sum_{j=1}^{K} t_{i j} \log \frac{t_{i j}}{s_{i j}}\).
2-2. PSOM : Probabilistic SOM clustering
propose a novel clustering called PSOM (Probabilistic SOM)
-
PSOM = extends CAH to include SOM
- combine \(\mathcal{L}_{\mathrm{CAH}}\) with \(\mathcal{L}_{\mathrm{S-SOM}}\) ( = Soft SOM loss )
- to get an interpretable representation
PSOM = SOM + soft cluster assignment
- probability that \(z_i\) belongs to cluster centroid \(\mu_j\) : \(s_{i j}=\frac{\left(1+ \mid \mid z_{i}-\mu_{j} \mid \mid ^{2} / \alpha\right)^{-\frac{\alpha+1}{2}}}{\sum_{j^{\prime}}\left(1+ \mid \mid z_{i}-\mu_{j^{\prime}} \mid \mid ^{2} / \alpha\right)^{-\frac{\alpha+1}{2}}}\).
Loss Function
\(\mathcal{L}_{\mathrm{PSOM}}=\mathcal{L}_{\mathrm{CAH}}+\beta \mathcal{L}_{\mathrm{S}-\mathrm{SOM}}\).
- \(\mathcal{L}_{\mathrm{S}-\mathrm{SOM}}=-\frac{1}{N} \sum_{i=1}^{N} \sum_{j=1}^{K} s_{i j} \sum_{e \in N(j)} \log s_{i e}=\sum_{z=1}^{Z}-\frac{1}{N} \sum_{i=1}^{N} \sum_{j=1}^{K} s_{i j} \log s_{i n_{z}(j)}\)>
2-3. DPSOM : VAE for representation learning
to increase expressivity of PSOM
-
apply clustering in the latent space of DEEP representation learning model
- non-linear mapping \(x_i \rightarrow z_i\) : by VAE
- learns prob distn \(q_{\phi}\left(z_{i} \mid x_{i}\right)\)
- parameterized using MVN, \(\left(\mu_{\phi}, \Sigma_{\phi}\right)=f_{\phi}\left(x_{i}\right)\)
- learns prob distn \(q_{\phi}\left(z_{i} \mid x_{i}\right)\)
- VAE loss ( = ELBO )
- \(\mathcal{L}_{\mathrm{VAE}}=\sum_{i=1}^{N}\left[-\mathbb{E}_{q_{\phi}\left(z \mid x_{i}\right)}\left(\log p_{\theta}\left(x_{i} \mid z\right)\right)+D_{K L}\left(q_{\phi}\left(z \mid x_{i}\right) \mid \mid p(z)\right)\right]\).
Loss Function
\(\mathcal{L}_{\mathrm{DPSOM}}=\gamma \mathcal{L}_{\mathrm{CAH}}+\beta \mathcal{L}_{\mathrm{S}-\mathrm{SOM}}+\mathcal{L}_{\mathrm{VAE}}\).
2-4. T-DPSOM : Extension to time series data
add temporal component
given set of \(N\) time series of length \(T\), \(\left\{x_{i, t}\right\}_{i=1, \ldots, N ; t=1, \ldots, T}\)
\(\rightarrow\) goal : learn INTERPRETABLE trajectories on the SOM grid
add additional loss term, which is similar to the smoothness loss in SOM-VAE ( + soft assignments )
- \(\mathcal{L}_{\text {smooth }}=-\frac{1}{N T} \sum_{i=1}^{N} \sum_{t=1}^{T} u_{i_{t}, i_{t+1}}\).
- where \(u_{i_{t}, i_{t+1}}=g\left(z_{i, t}, z_{i, t+1}\right)\) is the similarity between \(z_{i, t}\) and \(z_{i, t+1}\) using a Student’s t-distribution
- \(z_{i, t}\) refers to the embedding of time series \(x_{i}\) at time index \(t\).
- maximize the similarity between latent embeddings of adjacent time steps
one of main goals in t.s modeling : predicting future data points
- can be achieved by adding LSTM over latent embedding
- at each time step \(t\), predict prob distn \(p_{\omega}\left(z_{t+1} \mid z_{t}\right)\)
\(\rightarrow\) add prediction loss
- \(\mathcal{L}_{\text {pred }}=-\sum_{i=1}^{N} \sum_{t=1}^{T} \log p_{\omega}\left(z_{t+1} \mid z_{t}\right)\).
Loss Function
\(\mathcal{L}_{\mathrm{T}-\mathrm{DPSOM}}=\mathcal{L}_{\mathrm{DPSOM}}+\mathcal{L}_{\text {smooth }}+\mathcal{L}_{\text {pred }}\).
