TimeSiam: A Pre-Training Framework for Siamese Time-Series Modeling
( SimMTM 저자 )
https://arxiv.org/pdf/2402.02475.pdf
Contents
- Abstract
- Introduction
- TimeSiam
- Pre-training
- Fin-tuning
- Experiments
Abstract
Randomly masking TS or calculating series-wise similarity
\(\rightarrow\) Neglect inherent temporal correlations
Time-Siam
-
TS + Siamese Network
-
Pretrains siamese encoders to capture intrinsic temporal correlations between
randomly sampled (1) past and (2) current subseries
- Simple DA (e.g. masking)
- Benefit from diverse augmented subseries and learn internal time-dependent representations through a past-to-current reconstruction
- Learnable lineage embeddings
- To distinguish temporal distance between sampled series
- To foster the learning of diverse temporal correlations
- Experiments) Forecasting and Classification
- across 13 standard benchmarks in both intra- and cross-domain scenarios.
1. Introduction
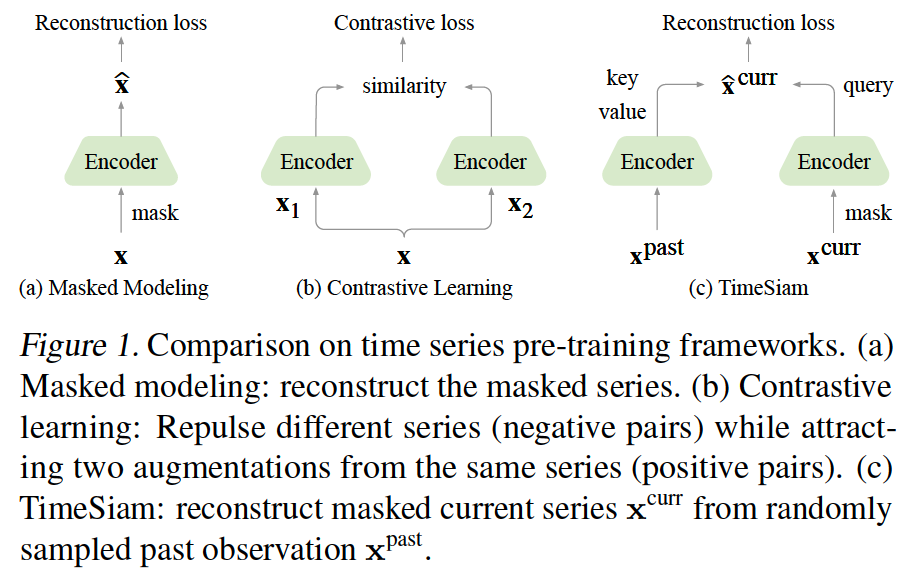
Two paradigms
- (1) Masked modeling
- Randomly masking a portion of time points will seriously distort vital temporal correlations of time series (Dong et al. (2023))
- (2) Contrastive learning
- Excels in instance-level representation learning
- Limitation: Reliance on augmentations to learn useful invariances (Xiao et al., 2020)
- May fail in capturing fine-grained temporal variations
Critical point of TS pre-training is optimizing encoders to accurately capture temporal correlations
TimeSiam
-
Simple yet effective self-supervised pre-training framework
-
[Figure 1-(c)] Sample pairs of subseries across different timestamps from the same TS
( = “Siamese subseries” )
- Leverages Siamese networks to capture correlations between temporally distanced subseries
-
Simple data augmentation
-
Not constrained by proximity information in TS
- Effectively model the correlation among distanced subseries
- empowers the model with a more thorough understanding of the whole TS
2. TimeSiam
Past-to-current reconstruction task with simple masked augmentation
( + Learnable lineage embeddings : to dynamically capture the disparity among different distanced subseries pairs )
(1) Pre-training
Two modules
- (1) Siamese subseries sampling
- (2) Siamese modeling
a) Siamese Subseries Sampling
- (Previous) Solely on modeling the individual series itself
- neglecting the inherent correlations among temporally related TS
- (TimeSiam) Focus on modeling temporal correlations of subseries across different timestamps
- capturing the intrinsic time-correlated information of TS
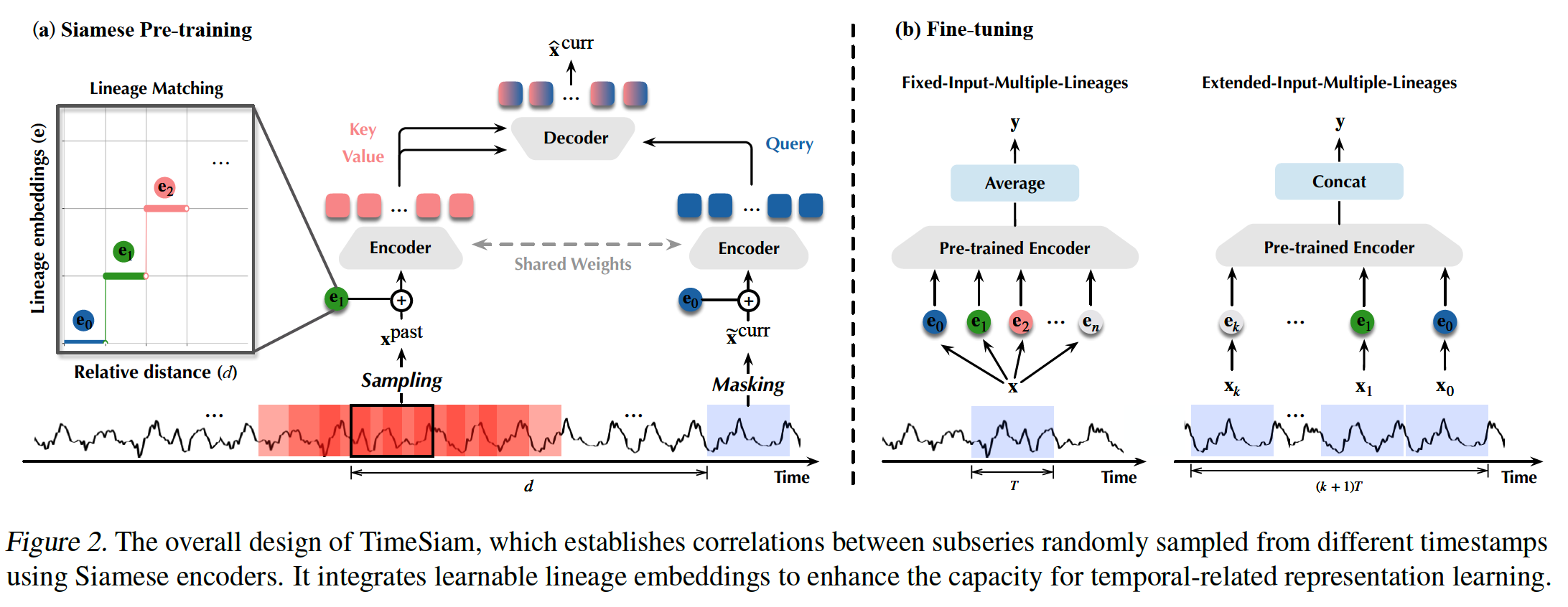
- “Siamese subseries” ( = Subseries pairs )
- By randomly sampling a past sample \(\mathrm{x}^{\text {past }}\) preceding the current sample \(\mathbf{x}^{\text {curr }}\) in the same TS
- Each contains \(T\) timestamps and \(C\) observed variables
- By randomly sampling a past sample \(\mathrm{x}^{\text {past }}\) preceding the current sample \(\mathbf{x}^{\text {curr }}\) in the same TS
- Goal: Constructing correlations and capturing temporal variations between these Siamese subseries
- Relative distance between the past and current subseries : \(d\)
- Simple masking augmentation
- To generate augmented current subseries \(\widetilde{\mathbf{x}}^{\text {curr }}\) that further improves the diversity and the disparity of Siamese subseries pairs
- \(\left(\mathrm{x}^{\text {past }}, \widetilde{\mathrm{x}}^{\text {curr }}\right)=\text { Mask-Augment }\left(\left(\mathrm{x}^{\text {past }}, \mathrm{x}^{\text {curr }}\right)\right)\).
b) Siamese Modeling
Lineage embeddings
-
Integrate learnable lineage embeddings during pre-training
-
to effectively capture the disparity among different Siamese pairs
-
enhance the model’s capacity to extract diverse temporal-related representations
-
-
Used to identify the temporal distance between Siamese subseries
-
\(N\) learnable lineage embeddings \(\left\{\mathbf{e}_i^{\text {lineage }}\right\}_{i=1}^N\)
- \(\mathbf{e}_i^{\text {lineage }} \in \mathbb{R}^{1 \times D}\),
Lineage embeddings for PAST & CURRENT
- For the past sample \(\mathrm{x}^{\text {past }}\)…
- apply the LineageMatching \((\cdot)\)
- to dynamically match a certain lineage embedding based on its temporal distance \(d\) to the current series
- For the current sample \(\widetilde{\mathbf{x}}^{\text {curr }}\)…
- use a special lineage embedding to represent a degeneration situation as \(d=0\)
\(\begin{aligned} \mathbf{e}_i^{\text {lineage }} & =\operatorname{LineageMatching}(d) \\ \mathbf{z}^{\text {past }} & =\operatorname{Embed}\left(\mathbf{x}^{\text {past }}\right) \oplus \mathbf{e}_i^{\text {lineage }} \\ \widetilde{\mathbf{z}}^{\text {curr }} & =\operatorname{Embed}\left(\widetilde{\mathbf{x}}^{\text {curr }}\right) \oplus \mathbf{e}_0^{\text {lineage }} \end{aligned}\).
- where \(\mathbf{e}_0^{\text {lineage }} \in \mathbb{R}^{1 \times D}\) is the specific embedding for current subseries
- \(\mathbf{z}^{\text {past }}, \widetilde{\mathbf{z}}^{\text {curr }} \in \mathbb{R}^{T \times D}\) .
Utilizes Siamese encoders
- can be instantiated as advanced TS models
- e.g. PatchTST (Nie et al., 2023) or iTransformer (Liu et al., 2024).
-
\(\mathbf{h}_e^{\text {past }}=\operatorname{Encoder}\left(\mathbf{z}^{\text {past }}\right)\).
- \(\widetilde{\mathbf{h}}_e^{\text {curr }}=\operatorname{Encoder}\left(\widetilde{\mathbf{z}}^{\text {curr }}\right)\).
where \(\mathbf{h}_e^{\text {past }}, \tilde{\mathbf{h}}_e^{\text {curr }} \in \mathbb{R}^{T \times D}\)
Past-to-current reconstruction task
- Use a decoder that integrates cross-attention and self-attention
- to incorporate past information into the current subseries for reconstruction
- \(\widetilde{\mathbf{h}}_e^{\text {curr }}\) : query
- \(\mathbf{h}_e^{\text {past }}\) : key and value
\(\mathbf{h}_d=\operatorname{Decoder}\left(\widetilde{\mathbf{h}}_e^{\text {curr }}, \mathbf{h}_e^{\text {past }}\right)\).
-
Generate the decoder representation of the current time subseries, denotes as \(\widehat{\mathbf{h}}_d\).
- \(\begin{aligned} \widehat{\mathbf{h}}_d & =\operatorname{LayerNorm}\left(\widetilde{\mathbf{h}}_e^{\text {curr }}+\operatorname{Cross-Attn}\left(\widetilde{\mathbf{h}}_e^{\text {curr }}, \mathbf{h}_e^{\text {past }}, \mathbf{h}_e^{\text {past }}\right)\right) \\ \mathbf{h}_d^{\prime} & =\operatorname{LayerNorm}\left(\widehat{\mathbf{h}}_d+\operatorname{Self-Attn}\left(\widehat{\mathbf{h}}_d, \widehat{\mathbf{h}}_d, \widehat{\mathbf{h}}_d\right)\right. \\ \mathbf{h}_d & =\operatorname{LayerNorm}\left(\mathbf{h}_d^{\prime}+\operatorname{FFN}\left(\mathbf{h}_d^{\prime}\right)\right) . \end{aligned}\).
- Output of the decoder \(\mathbf{h}_d \in \mathbb{R}^{T \times D}\)
Reconstruction:
- \(\widehat{\mathbf{x}}^{\text {curr }}=\operatorname{Projector}\left(\mathbf{h}_d\right)\).
- \(\mathcal{L}_{\text {reconstruction }}= \mid \mid \mathrm{x}^{\text {curr }}-\widehat{\mathbf{x}}^{\text {curr }} \mid \mid _2^2\).
(2) Fine-tuning
Can capture diverse temporal related representations under different lineage embeddings
Two types of fine-tuning paradigms
-
(1) Fixed input series setting
-
(2) Extended input series setting
Fixed-Input-Multiple-Lineages
-
[Standard] Generates only one type of representation
-
[TimeSiam] Pretrains Siamese encoders with diverse lineage embeddings to capture different distanced temporal correlations
\(\rightarrow\) Derive diverse representations with different lineages for the same input series
-
Enhances the diversity of extracted representations
-
\(\overline{\mathbf{h}}_e=\text { Average }\left(\mathbf{h}_{e, 0}, \mathbf{h}_{e, 1}, \ldots \mathbf{h}_{e, n}\right)\).
- Ensemble of a set of temporal representations derived from the same input series
- Input series \(\mathbf{x} \in \mathbb{R}^{T \times C}\),
- \(\mathbf{h}_{e, i}=\operatorname{Encoder}\left(\operatorname{Embed}(\mathbf{x}) \oplus \mathbf{e}_i^{\text {lineage }}\right)\).
Extended-Input-Multiple-Lineages
Model may receive longer records than the pre-training series
Can leverage multiple lineage embeddings trained under different temporal distanced pairs to different segments
\(\overline{\mathbf{h}}_e=\operatorname{Concat}\left(\mathbf{h}_{e, 0}, \mathbf{h}_{e, 1}, \ldots, \mathbf{h}_{e, k}\right)\).
- where \(\mathbf{h}_{e, i}=\operatorname{Encoder}\left(\operatorname{Embed}\left(\mathbf{x}_i\right) \oplus \mathbf{e}_{\text {LineageMatching }(i T)}^{\text {lineage }}\right)\).
- \(\overline{\mathbf{h}}_e \in \mathbb{R}^{(k+1) T \times D}\) denotes the extracted representation for extended input series.
3. Experiments
-
Forecasting & Classification
-
In- & Cross-domain setting
(1) Experimental Setup
a) Datasets
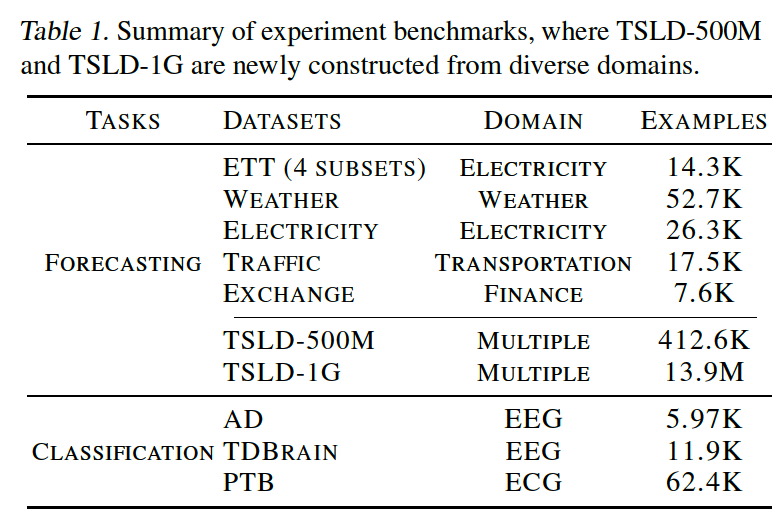
TSLD
- To further demonstrate the pre-training benefits under large and diverse data
- Constructed by merging time series datasets from multiple domains that are nonoverlapping with the other datasets
b) Backbone
-
iTransformerr & PatchTST for TS forecasting
-
TCN for TS classification
(2) Main Results
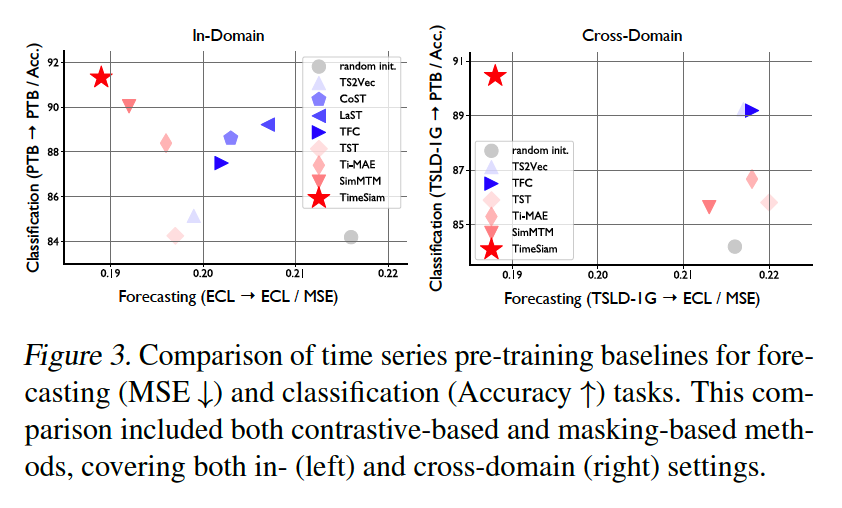
(3) Forecasting
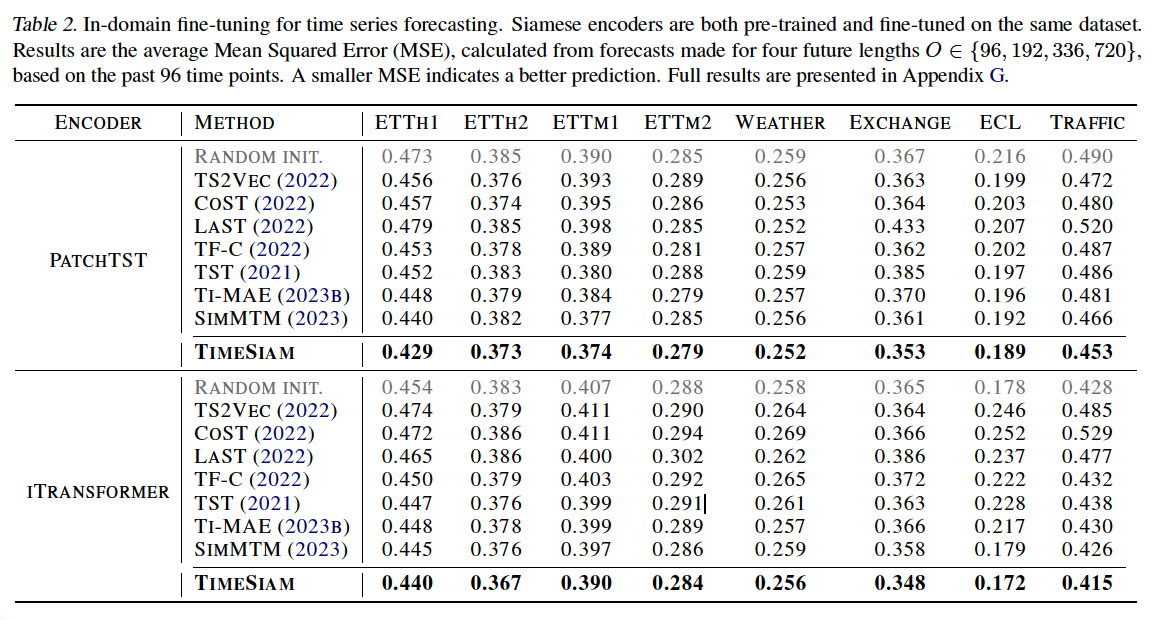
a) In-domain
b) Cross-domain
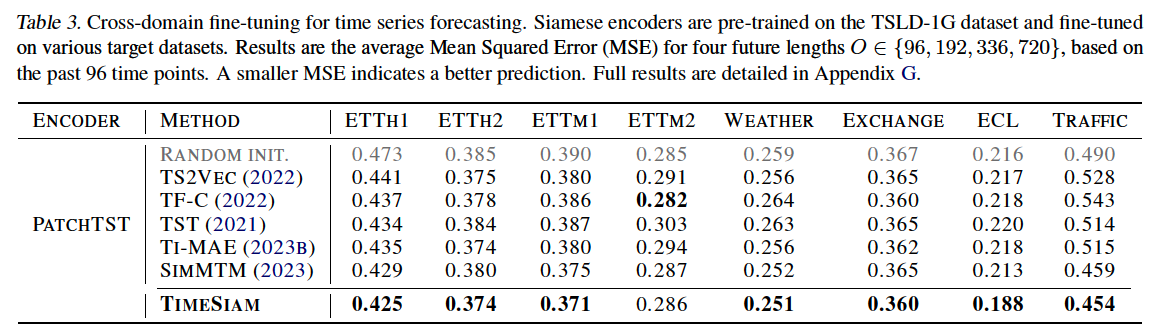
Use the TSLD-1G dataset
- Large-scale TS samples from diverse domains
- Even show superior performance compared to the in-domain scenario in some datasets
- ex) TSLD-1G → {ETTh1, ETTm1}.
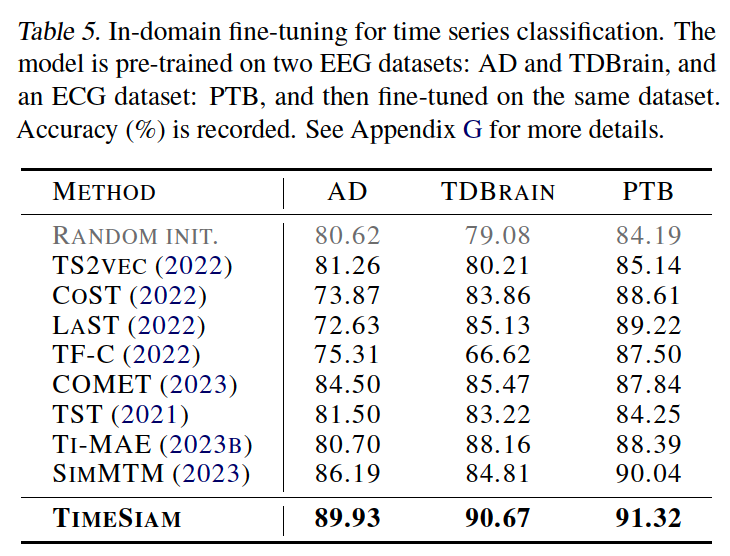
(4) Classification
a) In-domain
<br
b) Cross-domain
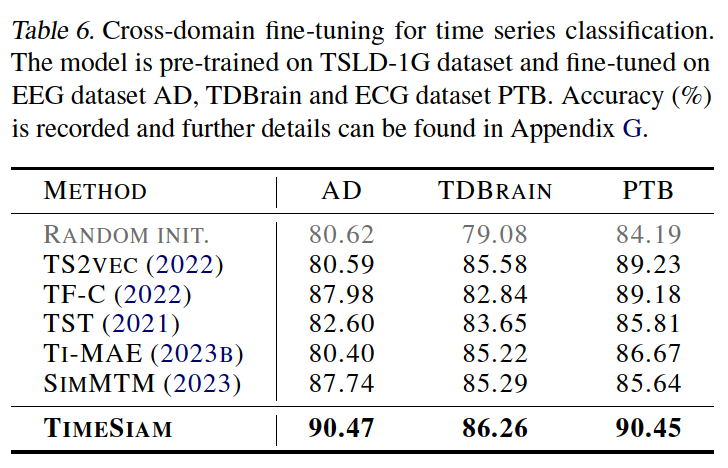
(5) Ablation Studies
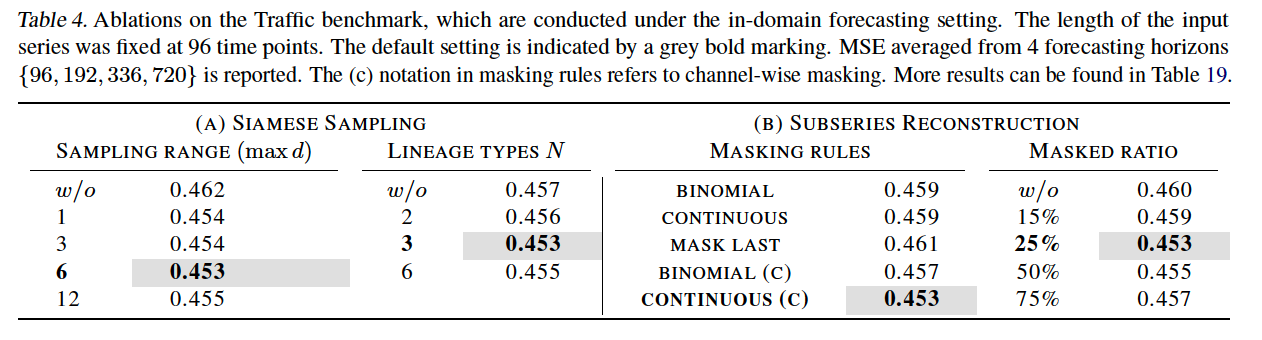
- (c) : channel-wise
(6) Analysis Experiment
a) Data Scale and Model Capacity
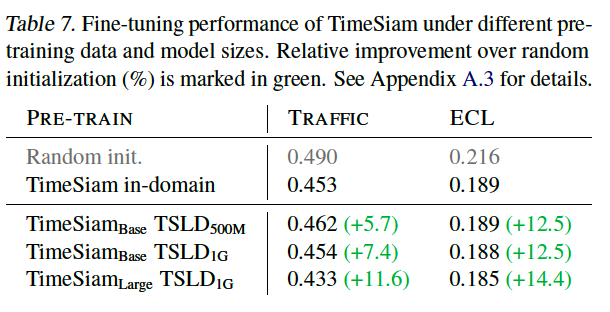
b) Adapt to Extended-Length Input
TimeSiam can natively adapt to longer inputs
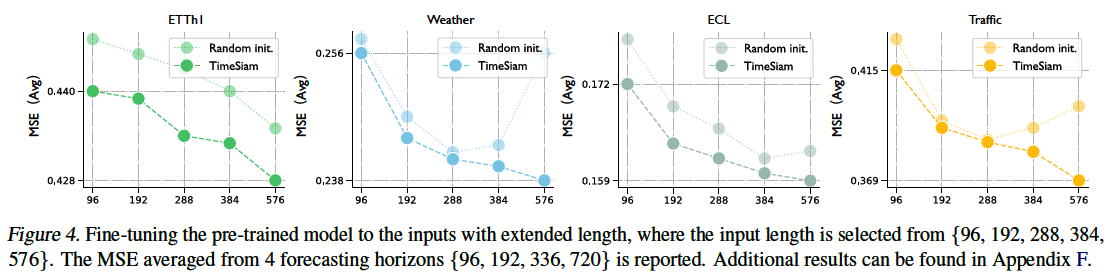
- [Standard] Degenerate under extended input length
c) Linear Probing
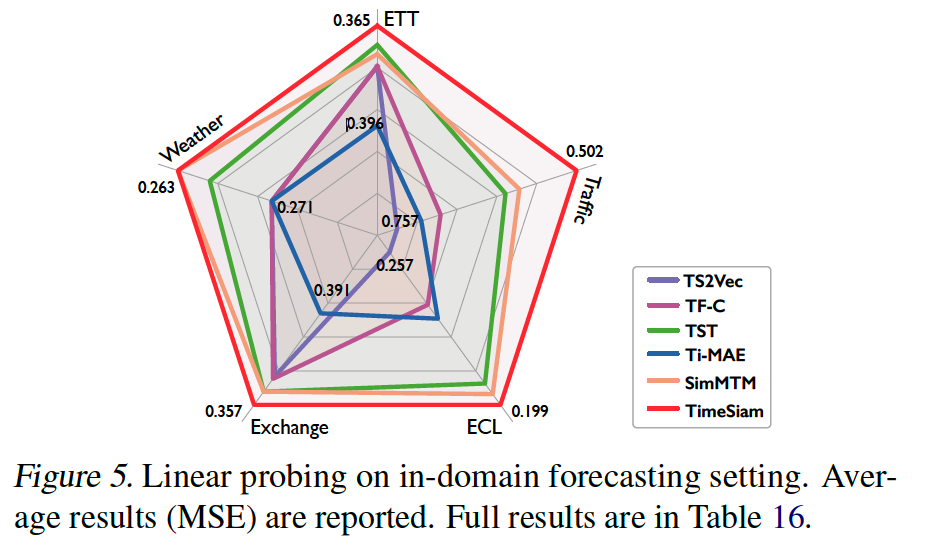
d) Embedding Effectiveness
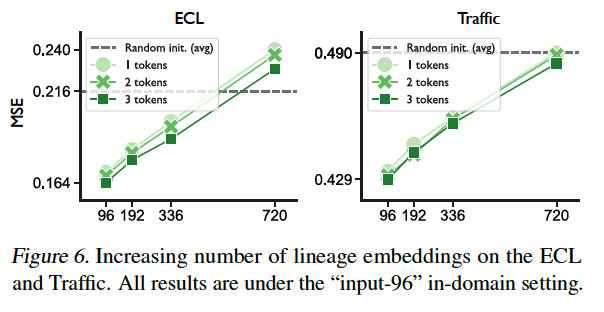
Advantages of employing varying numbers of lineage embeddings
\(\rightarrow\) Incorporation of lineage embeddings enhances prediction performance