[ 3. Node Embedding ]
( 참고 : CS224W: Machine Learning with Graphs )
Contents
- 3-1. Node Embeddings
- 3-2. Random Walk
- 3-3. Embedding Entire Graphs
3-1. Node Embeddings
1) Feature Representation ( Embedding )
Goal : create an…
- 1) efficient
- 2) task-independent feature learning
- 3) with graphs!
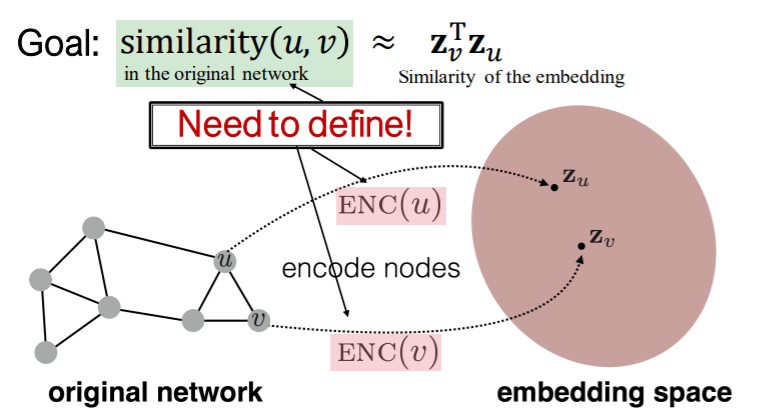
Similar in Network \(\approx\) Similar in Embedding Space
With these embeddings…solve many tasks!
- ex) node classification, link prediction….
Node Embedding algorithms
- ex) Deep Walk
2) Encoder & Decoder
Notation
- graph \(G\)
- vertex set \(V\)
- adjacency matrix \(A\) ( binary )
Goal : encode nodes, such that….
“similarity in embedding space \(\approx\) similarity in graph”
( \(\operatorname{similarity}(u, v) \approx \mathbf{z}_{v}^{\mathrm{T}} \mathbf{z}_{u}\) )
key point : how do we DEFINE SIMILARITY??
Process
-
step 1) node —-(encoder)——> embeddings
( \(\operatorname{ENC}(v)=\mathbf{z}_{v}\) )
-
step 2) define “node similarity function”
-
step 3) embedding—–(decoder)——> similarity score
-
step 4) optimization
( in a way that \(\operatorname{similarity}(u, v) \approx \mathbf{z}_{v}^{\mathrm{T}} \mathbf{z}_{u}\) )
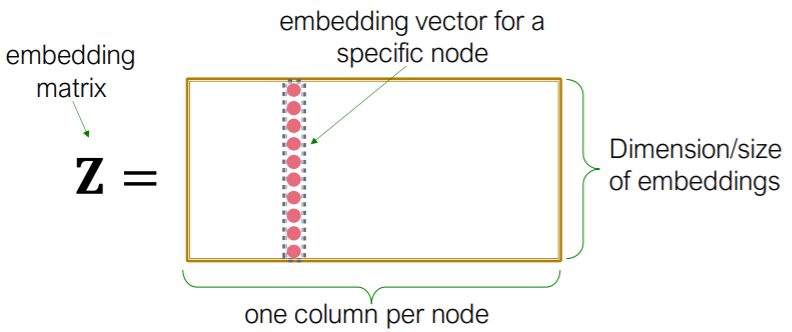
3) Shallow Encoding
- simplest ( just “embedding-lookup table” )
- \(\operatorname{ENC}(v)=\mathbf{z}_{v}=\mathbf{Z} \cdot v\).
- where \(\mathbf{Z} \in \mathbb{R}^{d \times \mid \mathcal{V} \mid }\) ( what we want to learn )
- \(v \in \mathbb{I}^{ \mid \mathcal{V} \mid }\) : indicator vector
- ex) Deep Walk, node2vec
4) How to define “Node Similarity”?
candidates : does/are 2 nodes…
- are linked?
- share neighbors?
- have similar roles in graph ( = structural roles )?
\(\rightarrow\) will learn “node similarity” that uses random walks!
Notes
- unsupervised / self-supervised tasks
- node labels (X), node features (X)
- these embeddings are “task independent”
3-2. Random Walk
1) Introduction
Notation
- \(z_u\) : embedding of \(u\)
- \(P(v \mid z_u)\) : (predicted) probability of visiting \(v\), starting from \(u\) ( on random walks )
- \(R\) : random walk strategy
Random Walk
- start from arbitrary node
- move to adjacent(neighbor) nodes RANDOMLY
- Random Walk : “sequence of points visited” in this way
- \(z^T_u z_v\) \(\approx\) probability that \(u\) & \(v\) co-occur on random walk
- nearby nodes : via \(N_R(u)\)
- neighborhood of \(u\), obtained by strategy \(R\)
2) Random Walk Embeddings
Random Walk Embeddings
- step 1) estimate \(P_R(v \mid u)\)
- step 2) optimize embeddings, to encode these random walk statistics
- cosine similarity \((z_i, z_j)\) \(\propto P_R(v \mid u)\)
Advantages of Random Walk
-
1) Expressivity
- captures both local & higher-order neighborhood info
-
2) Efficiency
-
do not consider ALL nodes
( only consider “PAIRS” that co-occur in random walk )
-
Feature Learning as Optimization
- goal :
- learn \(f(u) = z_u\)
- objective function :
- \(\max _{f} \sum_{u \in V} \log \mathrm{P}\left(N_{\mathrm{R}}(u) \mid \mathbf{z}_{u}\right)\).
3) Procedure
-
step 1) Run short fixed-length random walk
- starting node : \(u\)
- random walk strategy : \(R\)
-
step 2) collect \(N_R(u)\)
- ( = multiset of nodes visited )
-
step 3) optimize embedding
-
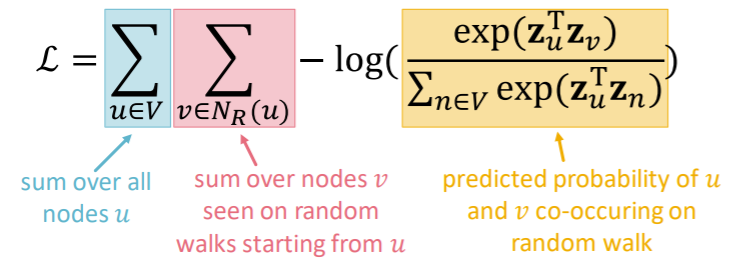
\(\max _{f} \sum_{u \in V} \log \mathrm{P}\left(N_{\mathrm{R}}(u) \mid \mathbf{z}_{u}\right)\).
( equivalently, \(\mathcal{L}=\sum_{u \in V} \sum_{v \in N_{R}(u)}-\log \left(P\left(v \mid \mathbf{z}_{u}\right)\right)\) )
-
\(P\left(v \mid \mathbf{z}_{u}\right)\) : parameterize as softmax
\(\rightarrow\) but TOO EXPENSIVE ( \(O(\mid V \mid^2)\) )… let’s approximate it
via NEGATIVE SAMPLING
-
4) Negative Sampling
\(\log \left(\frac{\exp \left(\mathbf{z}_{u}^{\mathrm{T}} \mathbf{z}_{v}\right)}{\sum_{n \in V} \exp \left(\mathbf{z}_{u}^{\mathrm{T}} \mathbf{z}_{n}\right)}\right) \approx \log \left(\sigma\left(\mathbf{z}_{u}^{\mathrm{T}} \mathbf{z}_{v}\right)\right)-\sum_{i=1}^{k} \log \left(\sigma\left(\mathbf{z}_{u}^{\mathrm{T}} \mathbf{z}_{n_{i}}\right)\right), n_{i} \sim P_{V}\).
-
do not use all nodes
only some “negative samples” \(n_i\)
( sampled not uniformly, but in a biased way )
-
biased way = “proportional to its degree”
-
appropriate \(k\)
- higher \(k\) : more robust & higher bias on negative events
- in practice, \(k=5 \sim 20\)
- optimize using SGD
5) Strategies of walking
- example) Deep Walk
- how to generalize?? “node2vec”
node2vec
- key point : biased 2nd order random walk to generate \(N_R(u)\)
- https://seunghan96.github.io/ne/ppt/4.node2vec/
3-3. Embedding Entire Graphs
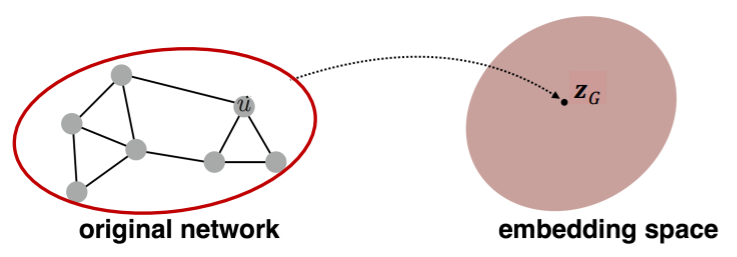
ex) classify toxic/non-toxic molecules
1) Idea 1 (simple)
- standard graph embedding
- sum the node embeddings in the graph
- \(\boldsymbol{z}_{\boldsymbol{G}}=\sum_{v \in G} Z_{v}\).
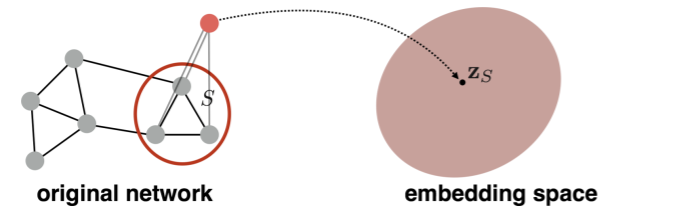
2) Idea 2
- use “virtual node”
- this node will represent the entire graph!
3) Idea 3 ( Anonymous Walk Embeddings )
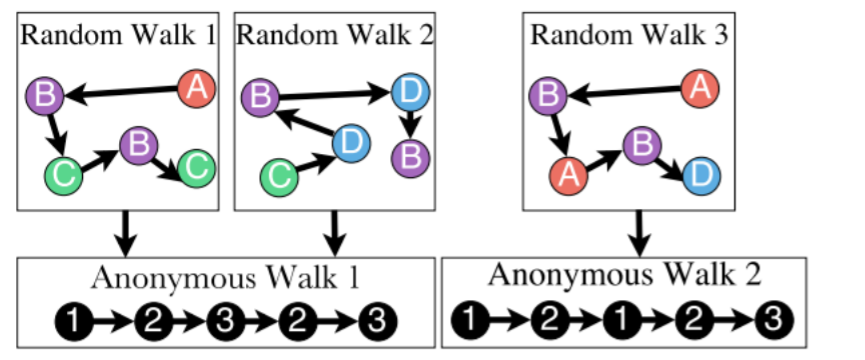
- do not consider which specific node it visisted!
- = agnostic to the identity
- = anonymous
- # of anonymous walks grows exponentially
- ex) \(l=3\)
- 5-dim representation
- 111,112,121,122,123
- \(Z_G[i]\) = prob of anonymous walk \(w_i\) in \(G\)
- sampling anonymous walks
- generate \(m\) random walks
- appropriate \(m\) ?
- \(m=\left[\frac{2}{\varepsilon^{2}}\left(\log \left(2^{\eta}-2\right)-\log (\delta)\right)\right]\).
- \(\eta\) : # of anonymous walks of length \(l\)
- \(m=\left[\frac{2}{\varepsilon^{2}}\left(\log \left(2^{\eta}-2\right)-\log (\delta)\right)\right]\).
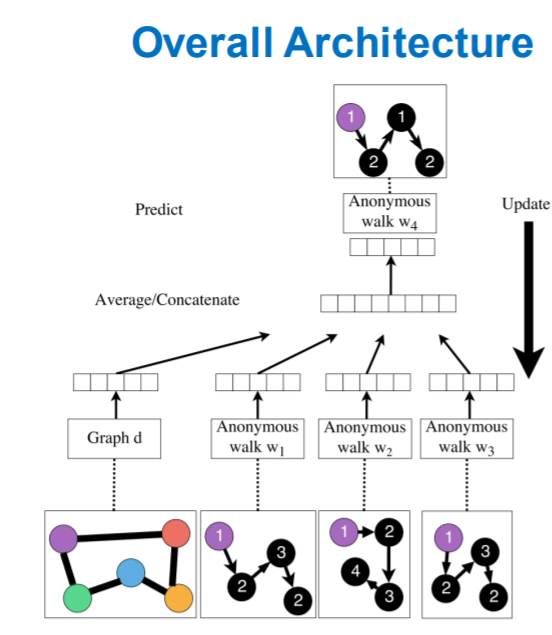
Learn Embeddings
- learn \(z_i\) ( = embedding of walk \(w_i\) )
-
learn \(Z_G\) ( = graph embedding, \(Z=\left\{z_{i}: i=1 \ldots \eta\right\}\) )
-
how to embed walks?
-
learn to predict walks, that co-occur in \(\Delta\) size window
-
\(\max \sum_{t=\Delta}^{T-\Delta} \log P\left(w_{t} \mid w_{t-\Delta}, \ldots, w_{t+\Delta}, z_{G}\right)\).
\(\rightarrow\) sum the objective, over ALL nodes
-
Process
- step 1) run \(T\) different random walks, of length \(l\)
- \(N_{R}(u)=\left\{w_{1}^{u}, w_{2}^{u} \ldots w_{T}^{u}\right\}\).
- step 2) optimize below
- \(\max _{\mathrm{Z}, \mathrm{d}} \frac{1}{T} \sum_{t=\Delta}^{T-\Delta} \log P\left(w_{t} \mid\left\{w_{t-\Delta}, \ldots, w_{t+\Delta} \mathbf{z}_{\boldsymbol{G}}\right\}\right)\).
- \(P\left(w_{t} \mid\left\{w_{t-\Delta}, \ldots, w_{t+\Delta}, z_{G}\right\}\right)=\frac{\exp \left(y\left(w_{t}\right)\right)}{\sum_{i=1}^{\eta} \exp \left(y\left(w_{i}\right)\right)}\).
- \(y\left(w_{t}\right)=b+U \cdot\left(\operatorname{cat}\left(\frac{1}{\Delta} \sum_{i=1}^{\Delta} \mathbf{z}_{i}, \mathbf{z}_{G}\right)\right)\).
- \(\operatorname{cat}\left(\frac{1}{\Delta} \sum_{i=1}^{\Delta} \mathbf{z}_{i}, \mathbf{z}_{G}\right)\) : avg of walk embeddings in the window, concatenated with graph embedding
- just treat $z_G$ like $z_i$, when optimizing!
- \(\max _{\mathrm{Z}, \mathrm{d}} \frac{1}{T} \sum_{t=\Delta}^{T-\Delta} \log P\left(w_{t} \mid\left\{w_{t-\Delta}, \ldots, w_{t+\Delta} \mathbf{z}_{\boldsymbol{G}}\right\}\right)\).
- step 3) obtain graph embedding \(Z_G\)
4) How to use embeddings
by using embeddings of nodes ( \(z_i\) )… we can solve…
- cluster/community detection
- cluster nodes
- node classification
- predict labels of nodes
- link prediction
- predict edges of 2 nodes
- how to use 2 node embeddings?
- 1) concatenate
- 2) Hadamard product
- 3) sum / average
- 4) distance
- graph classification
- get graph embedding \(Z_G\), by aggregating node embeddings
