[ CS224W - Colab 1 ]
( 참고 : CS224W: Machine Learning with Graphs )
import networkx as nx
1. Karate Club dataset
G = nx.karate_club_graph()
nx.draw(G, with_labels = True)
2. Statistics
2-1. Average Degree
def average_degree(num_edges, num_nodes):
avg_degree = (2*num_nodes) / num_edges
return round(avg_degree,0)
num_edges = G.number_of_edges()
num_nodes = G.number_of_nodes()
avg_degree = average_degree(num_edges, num_nodes) # 1.0
2-2. Average Clustering Coefficient
from itertools import combinations
def average_clustering_coefficient(G):
num_nodes = G.number_of_nodes()
edge_list = list(G.edges())
avg_cluster_coef = 0
for node_idx in range(num_nodes):
NB = list(G.neighbors(node_idx))
NB_comb = list(combinations(NB,2))
num_edges_NB = len(set(edge_list)&set(NB_comb))
if num_edges_NB>0:
cluster_coef = num_edges_NB/ len(NB_comb)
avg_cluster_coef +=cluster_coef
avg_cluster_coef /= num_nodes
return round(avg_cluster_coef,2)
avg_cluster_coef = average_clustering_coefficient(G) # 0.57
2-3. Page Rank
- 내용 참고 : https://wooono.tistory.com/189
num_iter만큼의 iteration이후, 나오게 되는 page rank 벡터
import numpy as np
def pagerank(G, beta, init_rank, num_iter):
pr = [init_rank]*G.number_of_nodes()
degrees = [val for (node, val) in G.degree()]
for _ in range(num_iter):
for node_idx in range(num_nodes):
pr[node_idx]=0
for neighbor_idx in list(G.neighbors(node_idx)):
pr[node_idx]+=pr[neighbor_idx]/degrees[neighbor_idx]
pr = np.array(pr)
pr*=beta
pr+=(1-beta)*(1/G.number_of_nodes())
return pr
beta = 0.8
init_rank = 1 / G.number_of_nodes()
num_iter = 10
pr1 = pagerank(G, beta, init_rank,num_iter)
Check with packages
pr2 = nx.pagerank(G,alpha=0.8)
pr2 = list(pr2.values())
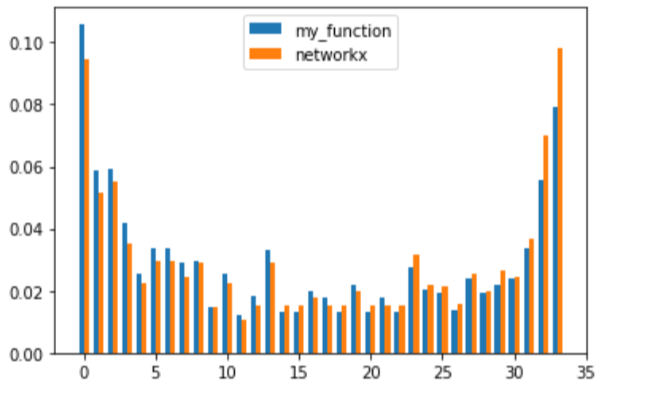
import matplotlib.pyplot as plt
x = np.arange(34)
width = 0.35
fig, ax = plt.subplots()
rects1 = ax.bar(x - width/2, pr1, width, label='my_function')
rects2 = ax.bar(x + width/2, pr2, width, label='networkx')
ax.legend()
2-4. Closeness Centrality
def closeness_centrality(G):
shortest_length = dict(nx.all_pairs_shortest_path_length(G))
cc_vec = []
for node_idx in G.nodes():
cc_vec.append(1/sum(shortest_length[node_idx].values()))
return cc_vec
closeness = closeness_centrality(G)
3. Graph to Tensor
3-1. transform to torch.LongTensor
def graph_to_edge_list(G):
return list(G.edges())
def edge_list_to_tensor(edge_list):
# method 1)
edge_index = torch.tensor(edge_list,dtype=torch.long)
'''
# method 2)
edge_index = torch.tensor(edge_list)
edge_index = edge_index.type(torch.long)
# method 3)
edge_index = torch.LongTensor(edge_list)
'''
return edge_index
pos_edge_list = graph_to_edge_list(G)
pos_edge_index = edge_list_to_tensor(pos_edge_list)
pos_edge_index
print("The pos_edge_index tensor has shape {}".format(pos_edge_index.shape))
print("The pos_edge_index tensor has sum value {}".format(torch.sum(pos_edge_index)))
print(pos_edge_index.dtype)
The pos_edge_index tensor has shape torch.Size([156, 2])
The pos_edge_index tensor has sum value 5070
torch.int64
4. Negative Sampling
import random
def sample_negative_edges(G, num_neg_samples,seed):
all_edges = list(combinations(list(G.nodes()),2))
pos_edges = list(G.edges())
neg_edges = list(set(all_edges)-set(pos_edges))
random.seed(seed)
neg_sam_idx = random.sample(range(len(neg_edges)),num_neg_samples)
neg_sam_edge_list = np.array(neg_edges)[neg_sam_idx]
return neg_sam_edge_list
neg_edge_list = sample_negative_edges(G, len(pos_edge_list),seed=960729)
neg_edge_index = edge_list_to_tensor(neg_edge_list)
5. Node Embedding
emb_sample = nn.Embedding(num_embeddings=4, embedding_dim=8)
print('Sample embedding layer: {}'.format(emb_sample))
Sample embedding layer: Embedding(4, 8)
Indexing
ids = torch.LongTensor([1, 3])
print(emb_sample(ids)) # size : 2x8
tensor([[-0.7282, 1.1633, -0.0091, -0.8425, 0.1374, 0.9386, -0.1860, -0.6446],
[ 0.4285, 1.4761, -1.7869, 1.6103, -0.7040, -0.1853, -0.9962, -0.8313]],
grad_fn=<EmbeddingBackward0>)
weight matrix
shape = emb_sample.weight.data.shape
print(shape)
torch.Size([4, 8])
Initialize with Uniform(0,1)
torch.manual_seed(1)
def create_node_emb(num_node=34, embedding_dim=16):
emb = nn.Embedding(num_node, embedding_dim)
emb.weight.data = torch.rand(num_node, embedding_dim)
return emb
emb = create_node_emb()
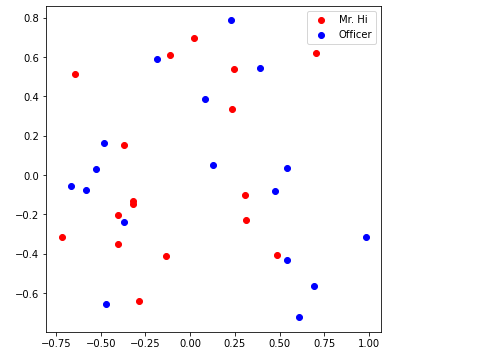
with sklearn’s PCA, can visualize as below!
visualize_emb(emb)
6. Training Embedding
from torch.optim import SGD
import torch.nn as nn
def accuracy(pred, label):
y_hat = (pred>0.5).type(torch.LongTensor)
acc = torch.mean((label==y_hat).type(torch.FloatTensor))
return acc
def train(G,emb,pos_edge_list, learning_rate=0.001,
num_iter_main=5,num_iter_sub=100):
total_iter=0
for _ in range(num_iter_main):
# (1) sample negative edge list
neg_edge_list = sample_negative_edges(G, len(pos_edge_list),seed=iter_main)
neg_edge_index = edge_list_to_tensor(neg_edge_list)
# (2) positive & negative embeddings
embed_pos = emb(pos_edge_index)
embed_neg = emb(neg_edge_index)
# (3) true values
pos_label = torch.ones(embed_pos.shape[0], )
neg_label = torch.zeros(embed_pos.shape[0], )
# (4) Run iterations
for _ in range(num_iter_sub):
opt.zero_grad()
y_pred_pos=sigmoid(torch.sum(embed_pos[:,0,:] * embed_pos[:,1,:], dim=-1))
y_pred_neg=sigmoid(torch.sum(embed_neg[:,0,:] * embed_neg[:,1,:], dim=-1))
y_pred=torch.cat([y_pred_pos,y_pred_neg])
y_true=torch.cat([pos_label,neg_label])
loss = loss_fn(y_pred,y_true)
loss.backward(retain_graph=True)
opt.step()
total_iter+=1
if total_iter%2000==0:
acc = accuracy(y_pred,y_true)
print('----------- Iteration : {} ---------'.format(total_iter))
print('Accuracy : ',acc.detach().numpy().round(3))
print('Loss : ',loss.detach().numpy().round(3))
sigmoid = nn.Sigmoid()
loss_fn = nn.BCELoss()
emb = create_node_emb(num_node=34, embedding_dim=16)
opt = SGD(emb.parameters(), lr=learning_rate, momentum=0.9)
train(G,emb,pos_edge_list, learning_rate=0.003,
num_iter_main=100,num_iter_sub=200)
----------- Iteration : 2000 ---------
Accuracy : 0.571
Loss : 0.634
----------- Iteration : 4000 ---------
Accuracy : 0.724
Loss : 0.494
..........
----------- Iteration : 18000 ---------
Accuracy : 0.872
Loss : 0.286
----------- Iteration : 20000 ---------
Accuracy : 0.897
Loss : 0.268
